文章目录
- install
- 使用org.Hs.egENSEMBL将Ensembl id convert to gene id
- org.Hs.egGENENAME 将Ensembl id convert to gene name
- org.Hs.egSYMBOL 将 gene symbol convert to gene id
- 我现在有一些ensembl id 如何转为 gene name
- 注意
- 你会遇到一些record不全的情况,gtf文件存在而org.Hs.eg.db不存在
install
# install
# if (!require("BiocManager", quietly = TRUE))
# install.packages("BiocManager")# BiocManager::install("AnnotationDbi")
# BiocManager::install("org.Hs.eg.db")# or install # wget https://www.bioconductor.org/packages/release/bioc/src/contrib/AnnotationDbi_1.62.2.tar.gz
# install.packages("/public/home/djs/software/AnnotationDbi_1.62.2.tar.gz", repos = NULL, type="source")
# wget https://www.bioconductor.org/packages/release/data/annotation/src/contrib/org.Hs.eg.db_3.17.0.tar.gz
# install.packages("/public/home/djs/software/org.Hs.eg.db_3.17.0.tar.gz", repos = NULL, type="source")
library(org.Hs.eg.db)help(package="org.Hs.eg.db")
Index:org.Hs.eg.db Bioconductor annotation data package
org.Hs.egACCNUM Map Entrez Gene identifiers to GenBank Accession Numbers
org.Hs.egALIAS2EG Map between Common Gene Symbol Identifiers and Entrez Gene
org.Hs.egCHR Map Entrez Gene IDs to Chromosomes
org.Hs.egCHRLENGTHS A named vector for the length of each of the chromosomes
org.Hs.egCHRLOC Entrez Gene IDs to Chromosomal Location
org.Hs.egENSEMBL Map Ensembl gene accession numbers with Entrez Gene identifiers
org.Hs.egENSEMBLPROT Map Ensembl protein acession numbers with Entrez Gene identifiers
org.Hs.egENSEMBLTRANS Map Ensembl transcript acession numbers with Entrez Gene identifiers
org.Hs.egENZYME Map between Entrez Gene IDs and Enzyme Commission (EC) Numbers
org.Hs.egGENENAME Map between Entrez Gene IDs and Genes
org.Hs.egGENETYPE Map between Entrez Gene Identifiers and Gene Type
org.Hs.egGO Maps between Entrez Gene IDs and Gene Ontology (GO) IDs
org.Hs.egMAP Map between Entrez Gene Identifiers and cytogenetic maps/bands
org.Hs.egMAPCOUNTS Number of mapped keys for the maps in package org.Hs.eg.db
org.Hs.egOMIM Map between Entrez Gene Identifiers and Mendelian Inheritance in Man (MIM) identifiers
org.Hs.egORGANISM The Organism for org.Hs.eg
org.Hs.egPATH Mappings between Entrez Gene identifiers and KEGG pathway identifiers
org.Hs.egPFAM Maps between Manufacturer Identifiers and PFAM Identifiers
org.Hs.egPMID Map between Entrez Gene Identifiers and PubMed Identifiers
org.Hs.egPROSITE Maps between Manufacturer Identifiers and PROSITE Identifiers
org.Hs.egREFSEQ Map between Entrez Gene Identifiers and RefSeq Identifiers
org.Hs.egSYMBOL Map between Entrez Gene Identifiers and Gene Symbols
org.Hs.egUNIPROT Map Uniprot accession numbers with Entrez Gene identifiers
org.Hs.eg_dbconn Collect information about the package annotation DB
使用org.Hs.egENSEMBL将Ensembl id convert to gene id
x <- org.Hs.egENSEMBL
# Get the entrez gene IDs that are mapped to an Ensembl ID
mapped_genes <- mappedkeys(x)
# Convert to a list
xx <- as.list(x[mapped_genes])xx[1:5] # entrez gene id 是list的索引名字,list的元素则是 ensembl id

org.Hs.egGENENAME 将Ensembl id convert to gene name
x <- org.Hs.egGENENAME
# Get the gene names that are mapped to an entrez gene identifier
mapped_genes <- mappedkeys(x)
# Convert to a list
xx <- as.list(x[mapped_genes])
org.Hs.egSYMBOL 将 gene symbol convert to gene id
x <- org.Hs.egSYMBOL
# Get the gene symbol that are mapped to an entrez gene identifiers
mapped_genes <- mappedkeys(x)
# Convert to a list
xx <- as.list(x[mapped_genes])# For the reverse map:
x <- org.Hs.egSYMBOL2EG
# Get the entrez gene identifiers that are mapped to a gene symbol
mapped_genes <- mappedkeys(x)
# Convert to a list
xx <- as.list(x[mapped_genes])

我现在有一些ensembl id 如何转为 gene name
# 将 ensembl id 单独拿出来
k <- keys(org.Hs.eg.db,keytype = "ENSEMBL")
# 然后根据 ensembl id 调出来entrez gene id 和 gene symbol
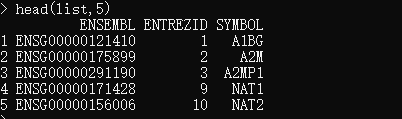
list <- select(org.Hs.eg.db,keys=k,columns = c("ENTREZID","SYMBOL"), keytype="ENSEMBL")# 或者使用你自己的 ensembl id 作为keys
list <- select(org.Hs.eg.db,keys=ID,columns = c("ENTREZID","SYMBOL"), keytype="ENSEMBL")head(list,5)
# 此处的 ensembl ID就是你个性化的id,我这里直接抽样得到然后用于演示
ID <- sample(list$ENSEMBL,10)
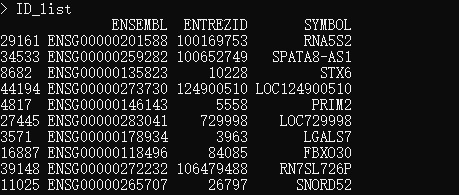
ID_list <- list[match(ID,list[,"ENSEMBL"]),]
ID_list
注意
这些ID对应关系随着不同数据库的升级和维护有可能出现前后不对应的情况。
同时这些ID 也不是一一对应的关系,可能存在一对多或者多对一的关系。
你会遇到一些record不全的情况,gtf文件存在而org.Hs.eg.db不存在
gtf存在 61544个基因

x <- org.Hs.egENSEMBLsum(is.na(unlist(as.list(x))))
[1] 105167
sum(!is.na(unlist(as.list(x))))
[1] 45727
# org.Hs.egENSEMBL 只有45727 个record
自己找个gtf文件然后提取信息再做转化吧
cat gencode.v40.annotation.gtf |awk 'BEGIN{FS=="\t"} $3~/gene/{print $0}' |cut -f 9 | cut -d ";" -f1,3 |cut -d " " -f2,4 |sed 's/\..*;//g' |sed 's/"//g' > ENSEMBL_TO_GENE.txt