Li, Yongge, et al. "CREaTor: zero-shot cis-regulatory pattern modeling with attention mechanisms." Genome Biology 24.1 (2023): 266.
2023年11月23日见刊
微信分享:Genome Biology | CREaTor: 零样本学习预测细胞类型特异的基因表达顺式调控模式
待解决问题:顺式-调控序列与靶基因连接 Linking cis-regulatory sequences to target genes
一、数据集:
RNA expression, DNase-seq, and ChIP-seq files were downloaded from ENCODE (ENCODE, by October 2021).
细胞类型:human tissues, primary cells, cell lines, and in vitro differentiated cells (1) with RNA-seq, DNase, CTCF ChIP-seq, H3K4me3 ChIP-seq, and H3K27ac ChIP-seq data available on ENCODE and (2) with complete cCRE information on SCREEN Registry V3.
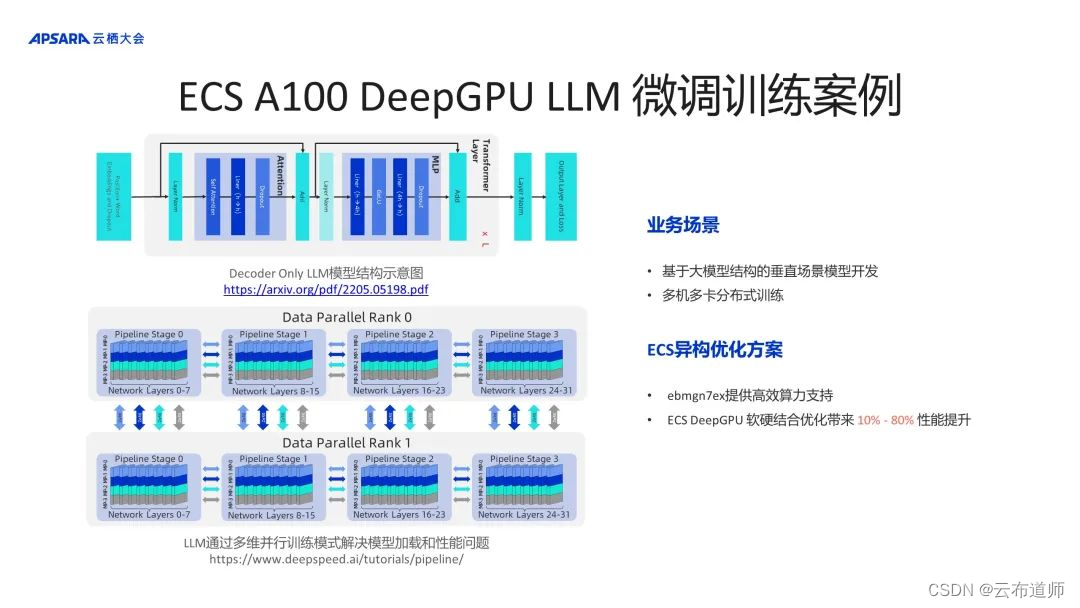
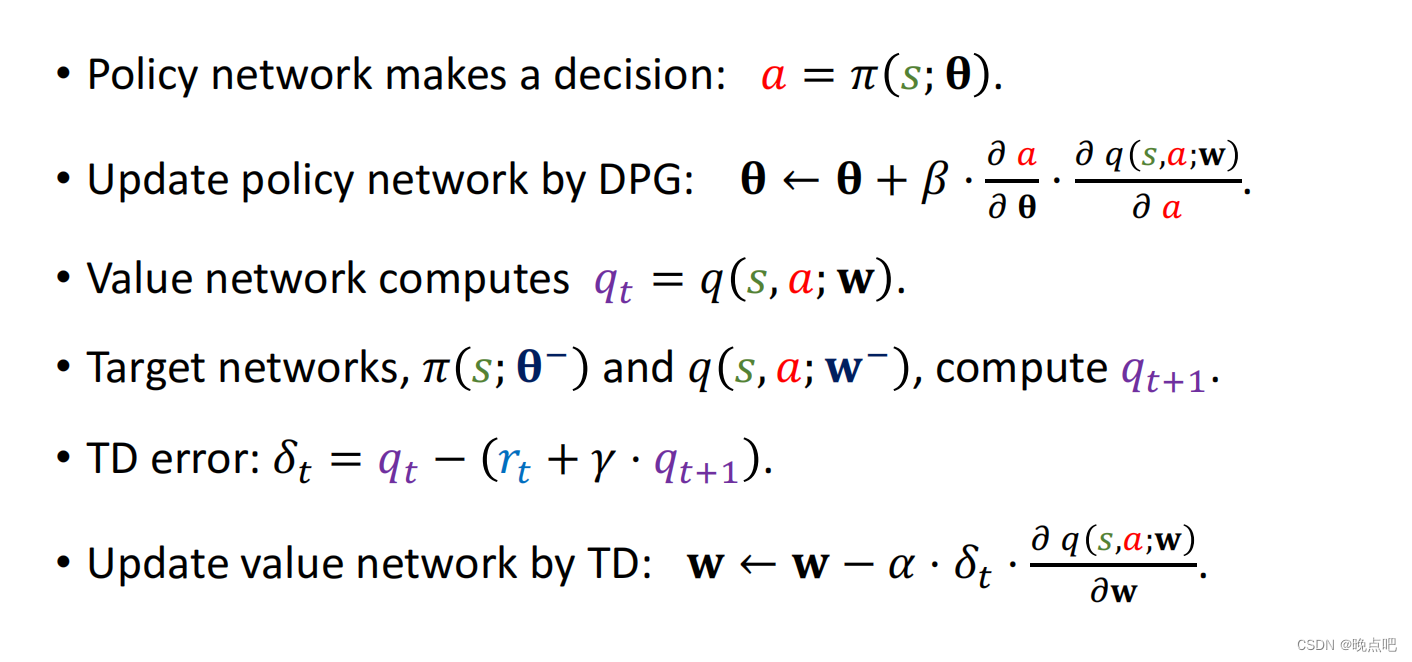
二、CREaTor 的架构
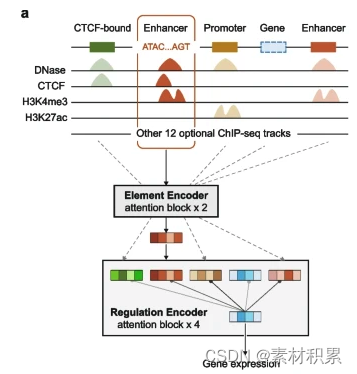
三、主要参考文献
1. Li, Yongge, et al. "Modeling cross-cell type cis-regulatory patterns via hierarchical deep neural network and gene expression prediction." bioRxiv (2023): 2023-03.
2. ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012 Sep 6;489(7414):57–74.
3. Fulco CP, Nasser J, Jones TR, Munson G, Bergman DT, Subramanian V, et al. Activity-by-contact model of enhancer–promoter regulation from thousands of CRISPR perturbations. Nat Genet. 2019 Dec;51(12):1664–9. 被引次数:605
4. Avsec Ž, Agarwal V, Visentin D, Ledsam JR, Grabska-Barwinska A, Taylor KR, et al. Effective gene expression prediction from sequence by integrating long-range interactions. Nat Methods. 2021 Oct;18(10):1196–203. 被引次数:406
5. Karbalayghareh A, Sahin M, Leslie CS. Chromatin interaction–aware gene regulatory modeling with graph attention networks. Genome Research. 2022 May 1;32(5):930-44. 被引次数 38






![yocto系列讲解[实战篇]93 - 添加Qtwebengine和Browser实例](https://img-blog.csdnimg.cn/f64e1f114d7045eaa57a680daec4395b.png)
![[Angular] 笔记 6:ngStyle](https://img-blog.csdnimg.cn/direct/81feef62cd9e403da92b957081298cd5.png)