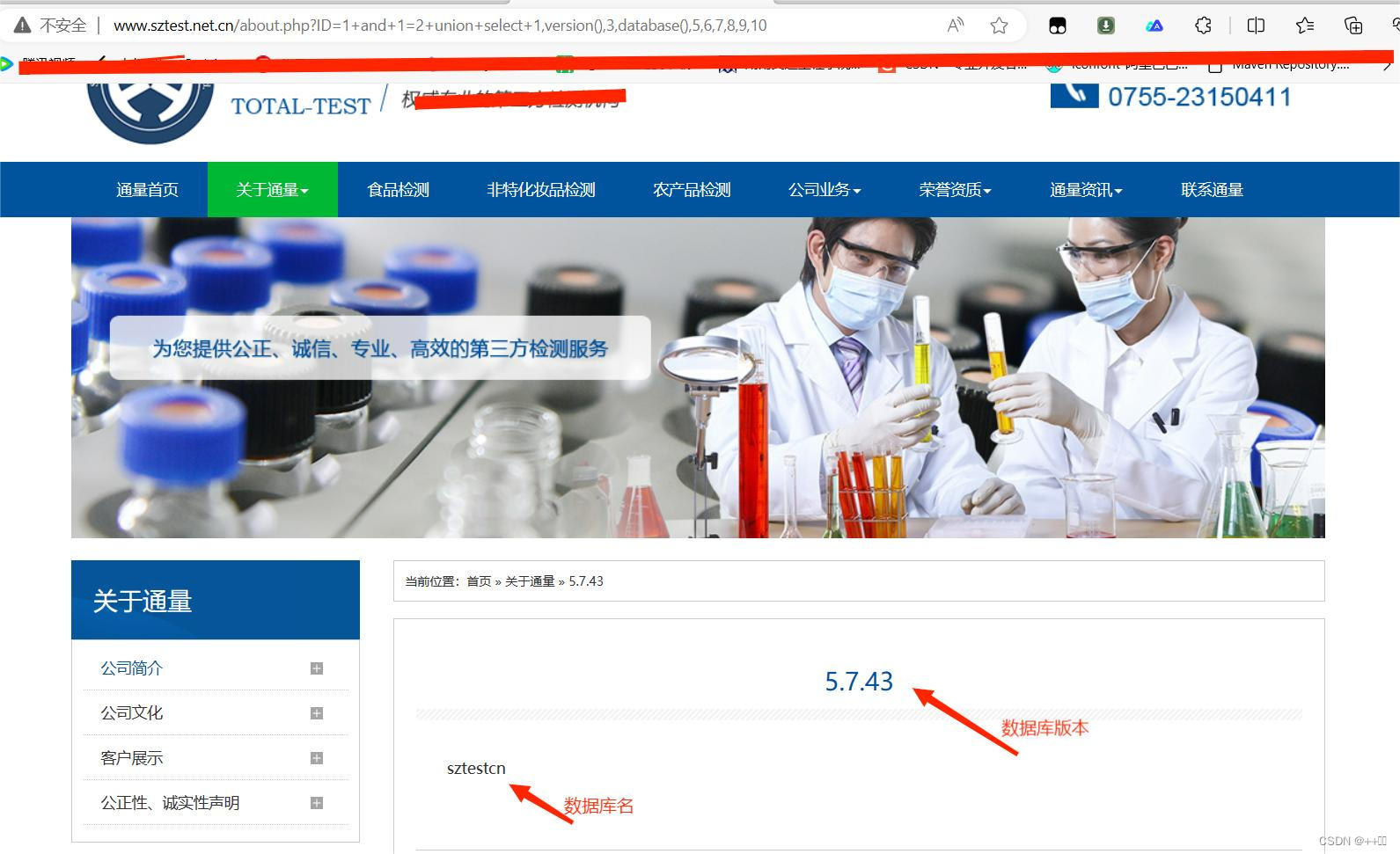
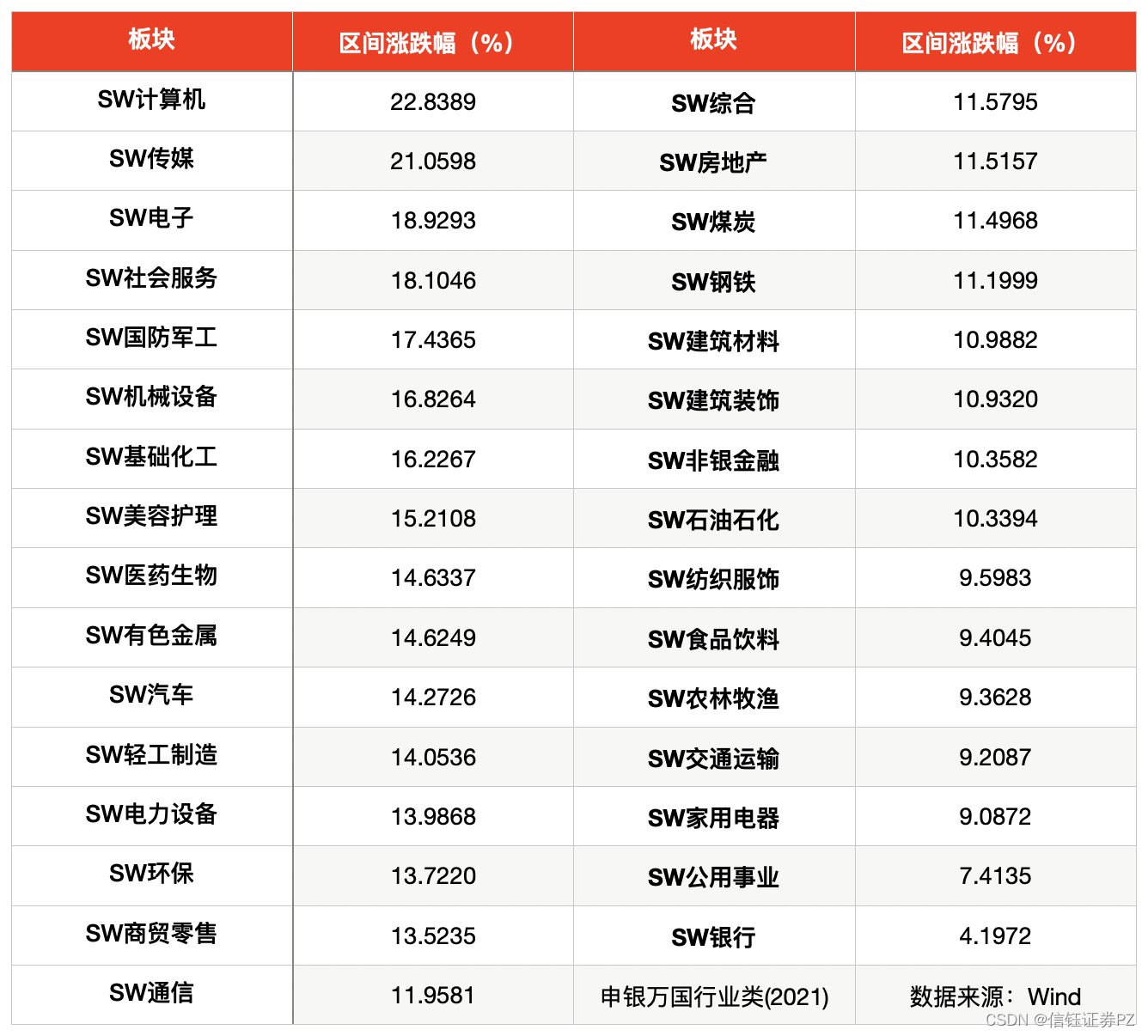
今天试一下安装PULpy
GitHub - WatsonLab/PULpy: Open prediction of Polysaccharide Utilisation Loci (PUL)
下载下面这个文件
https://github.com/WatsonLab/PULpy/blob/master/envs/PULpy.yaml
mkdir PULpy
cd PULpy
#将刚刚下的文件放到PULpy文件夹中
conda env create -f PULpy.yaml
source activate PULpy
#安装数据库
mkdir pfam_data && cd pfam_data
wget ftp://ftp.ebi.ac.uk/pub/databases/Pfam/current_release/Pfam-A.hmm.gz
wget ftp://ftp.ebi.ac.uk/pub/databases/Pfam/current_release/Pfam-A.hmm.dat.gz
wget ftp://ftp.ebi.ac.uk/pub/databases/Pfam/current_release/active_site.dat.gz
gunzip Pfam-A.hmm.gz Pfam-A.hmm.dat.gz active_site.dat.gz
hmmpress Pfam-A.hmm
cd ..
#接下来安装dbcan,但是我已经有了,待会看看能不能直接用
#这是官方让下载的
mkdir dbcan_data && cd dbcan_data
wget http://bcb.unl.edu/dbCAN2/download/Databases/dbCAN-old@UGA/hmmscan-parser.sh
wget http://bcb.unl.edu/dbCAN2/download/Databases/dbCAN-old@UGA/dbCAN-fam-HMMs.txt
hmmpress dbCAN-fam-HMMs.txt
chmod 755 hmmscan-parser.sh
cd ..
####
#如果你已经安装run_dbcan4,hmmscan-parser.sh文件在
~/miniconda3/envs/run_dbcan/bin
#数据库在这
https://bcb.unl.edu/dbCAN2/download/Databases/V12/dbCAN-HMMdb-V12.txt
复制github代码
git clone https://github.com/WatsonLab/PULpy修改config.json

chmod -R 755 scripts
mkdir genomes/
#将需要用的基因组放到上面文件夹中,并修改成以_genomic.fna.gz结尾
snakemake --use-conda --cores 90