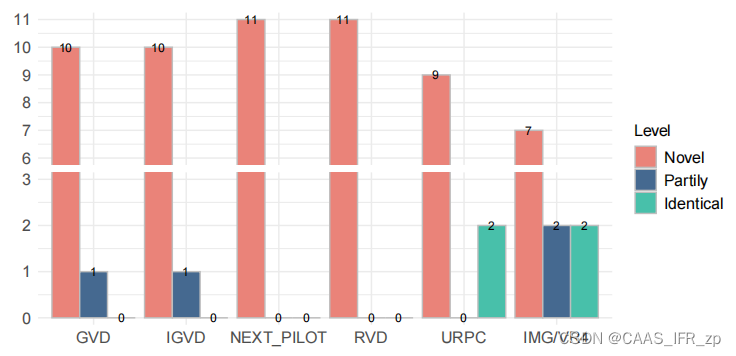
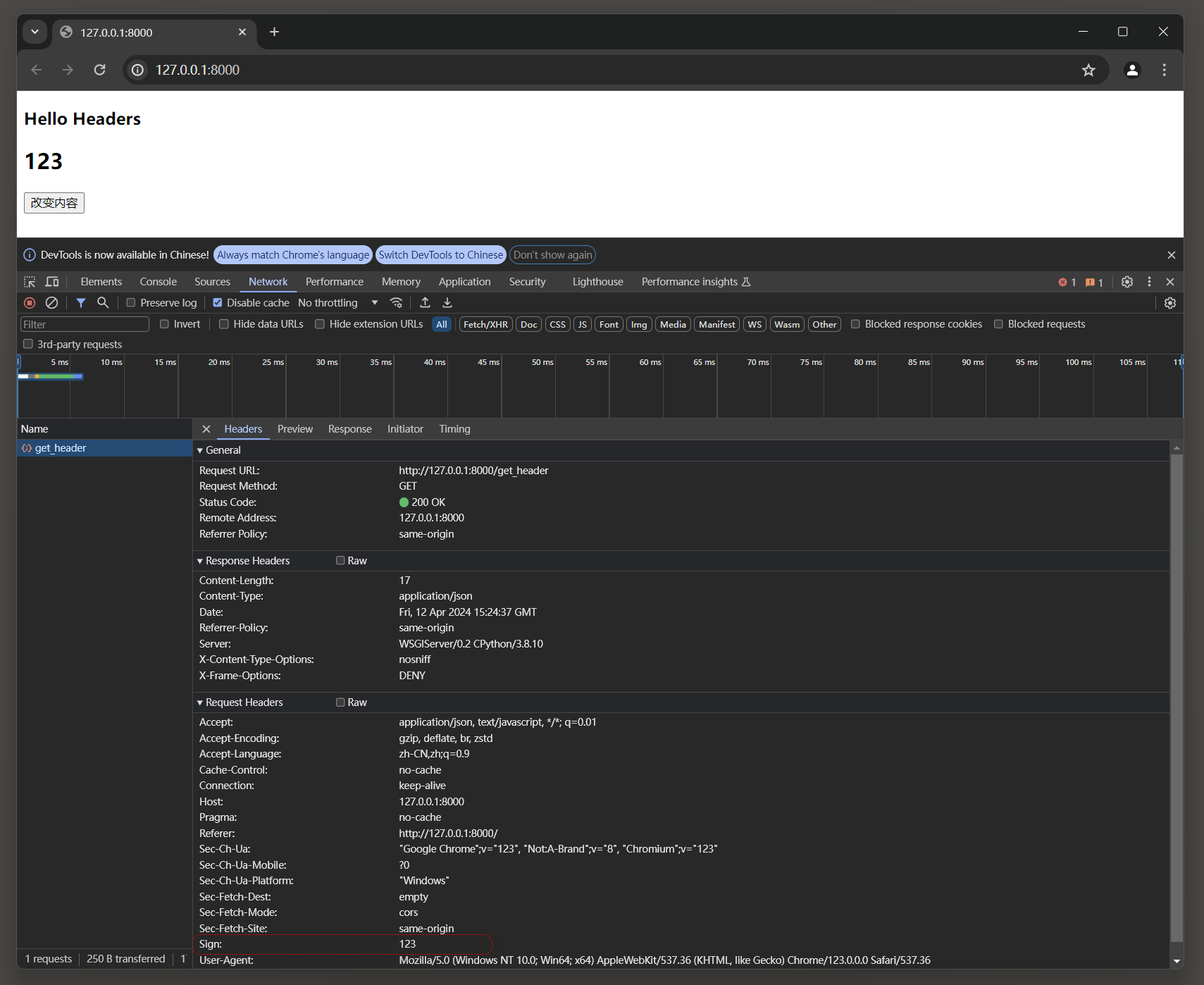
输入文件实例(存为csv格式)

library(ggplot2)
library(ggbreak)# 从CSV文件中读取数据
setwd("C:/Users/fordata/Desktop/研究生/第二个想法(16s肠型+宏基因组功能)/第二篇病毒组/result/otherDB")
data <- read.csv("fetal.csv")# 自定义颜色
my_colors <- c("Novel" = "#EA8379", "Partily" = "#456990", "Identical" = "#48C0AA")
data$DB <- factor(data$DB, levels = c("GVD", "IGVD", "NEXT_PILOT", "RVD", "URPC", "IMG/VR4"))
# 创建分组的柱状图
p <- ggplot(data, aes(fill=factor(Level, levels=c("Novel", "Partily", "Identical")), y=Count, x=DB)) + geom_bar(position="dodge", stat="identity", colour="gray", width=0.9) + scale_fill_manual(values=my_colors, name="Level") +theme_minimal() +theme(axis.text.x = element_text(size=12),axis.text.y = element_text(size=12),legend.text = element_text(size=12),legend.title = element_text(size=12)) + labs(x="", y="") +scale_y_break(c(3, 6), scales="free") # 截断,使得长的柱子不要太长# 在每个柱子上面添加数据标签
p <- p + geom_text(aes(label=Count), vjust=0.5, position=position_dodge(0.9), size=3)# 保存图像,并设置图像的宽度和高度
ggsave("2my_plot.pdf", plot = p, width = 8, height = 4)