4.1由于决策树只在样本同属于一类或者所有特征值都用完或缺失时生成叶节点,同一节点的样本,在路径上的特征值都相同,而训练集中又没有冲突数据,所以必定存在训练误差为0的决策树
4.2使用最小训练误差会导致过拟合,使得学习模型泛化能力下降。
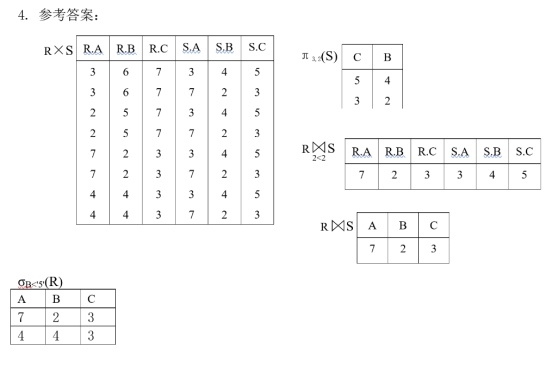
4.3
青绿0,乌黑1,浅白2
蜷缩0,稍蜷1,硬挺2
浊响0,沉闷1,清脆2
清晰0,烧糊1,模胡2
凹陷0,稍凹1,平坦2
硬滑0,软粘1
import numpy as np
import treePlotter
np.random.seed(100)
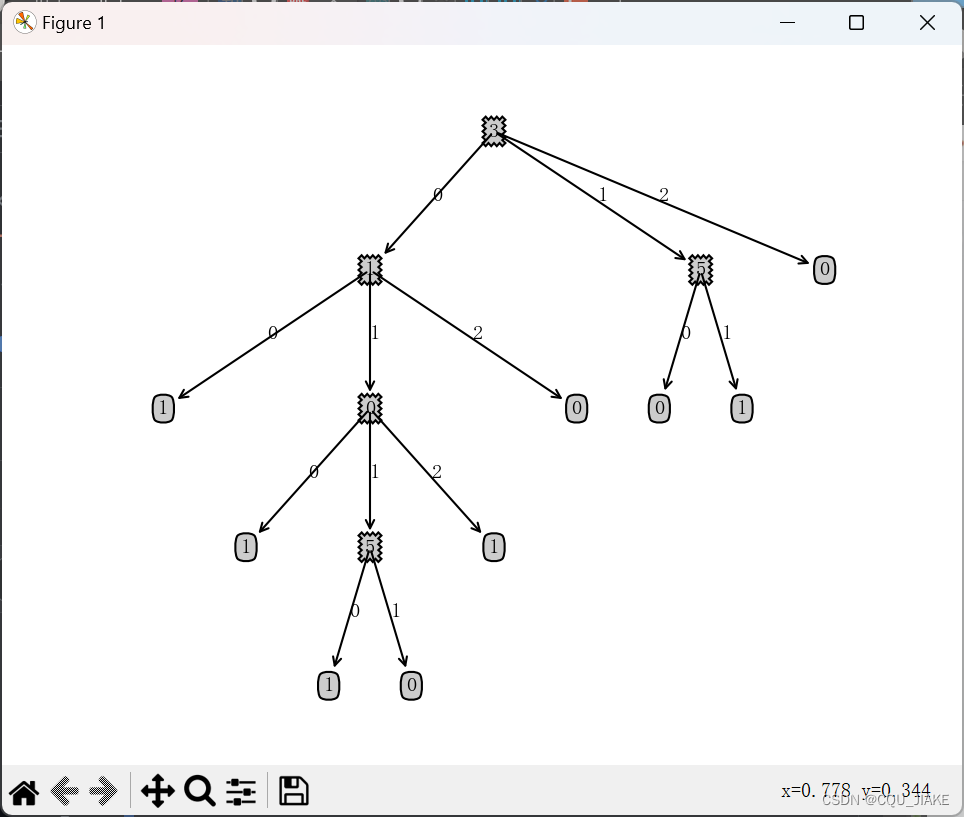
class DecisionTreeClassifier:def __init__(self,tree_type='ID3',k_classes=2):self.tree_type=tree_typeself.k_classes=k_classesif tree_type=='ID3':self.gain_func=self.Gainelif tree_type=='CART':self.gain_func=self.GiniIndexelif tree_type=='C45':self.gain_func=self.GainRatioelse:raise ValueError('must be ID3 or CART or C45')self.tree=Nonedef fit(self,X,y):D={}D['X']=XD['y']=yA=np.arange(X.shape[1])aVs={}for a in A:aVs[a]=np.unique(X[:,a])self.tree=self.TreeGenerate(D,A,aVs)def predict(self,X):if self.tree is None:raise RuntimeError('cant predict before fit')y_pred=[]for i in range(X.shape[0]):tree = self.treex=X[i]while True:if not isinstance(tree,dict):y_pred.append(tree)breaka=list(tree.keys())[0]tree=tree[a]if isinstance(tree,dict):val = x[a]tree = tree[val]else:y_pred.append(tree)breakreturn np.array(y_pred)def TreeGenerate(self,D,A,aVs):X=D['X']y=D['y']# 情形1unique_classes=np.unique(y)if len(unique_classes)==1:return unique_classes[0]flag=Truefor a in A:if(len(np.unique(X[:,a]))>1):flag=Falsebreak# 情形2if flag:return np.argmax(np.bincount(y))gains=np.zeros((len(A),))if self.tree_type=='C45':gains=np.zeros((len(A),2))for i in range(len(A)):gains[i]=self.gain_func(D,A[i])#print(gains)subA=Noneif self.tree_type=='CART':a_best=A[np.argmin(gains)]subA=np.delete(A,np.argmin(gains))elif self.tree_type=='ID3':a_best=A[np.argmax(gains)]subA=np.delete(A,np.argmax(gains))elif self.tree_type=='C45':gain_mean=np.mean(gains[:,0])higher_than_mean_indices=np.where(gains[:,0]>=gain_mean)higher_than_mean=gains[higher_than_mean_indices,1][0]index=higher_than_mean_indices[0][np.argmax(higher_than_mean)]a_best=A[index]subA=np.delete(A,index)tree={a_best:{}}for av in aVs[a_best]:indices=np.where(X[:,a_best]==av)Dv={}Dv['X']=X[indices]Dv['y']=y[indices]if len(Dv['y'])==0:tree[a_best][av]=np.argmax(np.bincount(y))else:tree[a_best][av]=self.TreeGenerate(Dv,subA,aVs)return tree@classmethoddef Ent(cls,D):y=D['y']bin_count=np.bincount(y)total=len(y)ent=0.for k in range(len(bin_count)):p_k=bin_count[k]/totalif p_k!=0:ent+=p_k*np.log2(p_k)return -ent@classmethoddef Gain(cls,D,a):X=D['X']y=D['y']aV=np.unique(X[:,a])sum=0.for v in range(len(aV)):Dv={}indices=np.where(X[:,a]==aV[v])Dv['X']=X[indices]Dv['y']=y[indices]ent=cls.Ent(Dv)sum+=(len(Dv['y'])/len(y)*ent)gain=cls.Ent(D)-sumreturn gain@classmethoddef Gini(cls,D):y = D['y']bin_count = np.bincount(y)total = len(y)ent = 0.for k in range(len(bin_count)):p_k = bin_count[k] / totalent+=p_k**2return 1-ent@classmethoddef GiniIndex(cls,D,a):X = D['X']y = D['y']aV = np.unique(X[:, a])sum = 0.for v in range(len(aV)):Dv = {}indices = np.where(X[:, a] == aV[v])Dv['X'] = X[indices]Dv['y'] = y[indices]ent = cls.Gini(Dv)sum += (len(Dv['y']) / len(y) * ent)gain = sumreturn gain@classmethoddef GainRatio(cls,D,a):X = D['X']y = D['y']aV = np.unique(X[:, a])sum = 0.intrinsic_value=0.for v in range(len(aV)):Dv = {}indices = np.where(X[:, a] == aV[v])Dv['X'] = X[indices]Dv['y'] = y[indices]ent = cls.Ent(Dv)sum += (len(Dv['y']) / len(y) * ent)intrinsic_value+=(len(Dv['y'])/len(y))*np.log2(len(Dv['y'])/len(y))gain = cls.Ent(D) - sumintrinsic_value=-intrinsic_valuegain_ratio=gain/intrinsic_valuereturn np.array([gain,gain_ratio])if __name__=='__main__':watermelon_data = np.array([[0, 0, 0, 0, 0, 0], [1, 0, 1, 0, 0, 0],[1, 0, 0, 0, 0, 0], [0, 0, 1, 0, 0, 0],[2, 0, 0, 0, 0, 0], [0, 1, 0, 0, 1, 1],[1, 1, 0, 1, 1, 1], [1, 1, 0, 0, 1, 0],[1, 1, 1, 1, 1, 0], [0, 2, 2, 0, 2, 1],[2, 2, 2, 2, 2, 0], [2, 0, 0, 2, 2, 1],[0, 1, 0, 1, 0, 0], [2, 1, 1, 1, 0, 0],[1, 1, 0, 0, 1, 1], [2, 0, 0, 2, 2, 0],[0, 0, 1, 1, 1, 0]])label = np.array([1, 1, 1, 1, 1, 1, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0])X_test=np.array([[0, 0, 1, 0, 0, 0], [1, 0, 1, 0, 0, 0],[1, 1, 0, 1, 1, 0], [1, 0, 1, 1, 1, 0],[1, 1, 0, 0, 1, 1], [2, 0, 0, 2, 2, 0],[0, 0, 1, 1, 1, 0]])decision_clf=DecisionTreeClassifier(tree_type='ID3')decision_clf.fit(watermelon_data,label)print(decision_clf.tree)treePlotter.createPlot(decision_clf.tree)y_pred=decision_clf.predict(X_test)print('y_pred:',y_pred)import matplotlib.pyplot as plt
from pylab import mpl
mpl.rcParams['font.sans-serif'] = ['FangSong']
mpl.rcParams['axes.unicode_minus'] = False
decisionNode = dict(boxstyle="sawtooth", fc="0.8")
leafNode = dict(boxstyle="round4", fc="0.8")
arrow_args = dict(arrowstyle="<-")def plotNode(nodeTxt, centerPt, parentPt, nodeType):createPlot.ax1.annotate(nodeTxt, xy=parentPt, xycoords='axes fraction', \xytext=centerPt, textcoords='axes fraction', \va="center", ha="center", bbox=nodeType, arrowprops=arrow_args)def getNumLeafs(myTree):numLeafs = 0firstStr = list(myTree.keys())[0]secondDict = myTree[firstStr]for key in secondDict.keys():if type(secondDict[key]).__name__ == 'dict':numLeafs += getNumLeafs(secondDict[key])else:numLeafs += 1return numLeafsdef getTreeDepth(myTree):maxDepth = 0firstStr = list(myTree.keys())[0]secondDict = myTree[firstStr]for key in secondDict.keys():if type(secondDict[key]).__name__ == 'dict':thisDepth = getTreeDepth(secondDict[key]) + 1else:thisDepth = 1if thisDepth > maxDepth:maxDepth = thisDepthreturn maxDepthdef plotMidText(cntrPt, parentPt, txtString):xMid = (parentPt[0] - cntrPt[0]) / 2.0 + cntrPt[0]yMid = (parentPt[1] - cntrPt[1]) / 2.0 + cntrPt[1]createPlot.ax1.text(xMid, yMid, txtString)def plotTree(myTree, parentPt, nodeTxt):numLeafs = getNumLeafs(myTree)depth = getTreeDepth(myTree)firstStr = list(myTree.keys())[0]cntrPt = (plotTree.xOff + (1.0 + float(numLeafs)) / 2.0 / plotTree.totalw, plotTree.yOff)plotMidText(cntrPt, parentPt, nodeTxt)plotNode(firstStr, cntrPt, parentPt, decisionNode)secondDict = myTree[firstStr]plotTree.yOff = plotTree.yOff - 1.0 / plotTree.totalDfor key in secondDict.keys():if type(secondDict[key]).__name__ == 'dict':plotTree(secondDict[key], cntrPt, str(key))else:plotTree.xOff = plotTree.xOff + 1.0 / plotTree.totalwplotNode(secondDict[key], (plotTree.xOff, plotTree.yOff), cntrPt, leafNode)plotMidText((plotTree.xOff, plotTree.yOff), cntrPt, str(key))plotTree.yOff = plotTree.yOff + 1.0 / plotTree.totalDdef createPlot(inTree):fig = plt.figure(1, facecolor='white')fig.clf()axprops = dict(xticks=[], yticks=[])createPlot.ax1 = plt.subplot(111, frameon=False, **axprops)plotTree.totalw = float(getNumLeafs(inTree))plotTree.totalD = float(getTreeDepth(inTree))plotTree.xOff = -0.5 / plotTree.totalwplotTree.yOff = 1.0plotTree(inTree, (0.5, 1.0), '')plt.show()4.5
import numpy as np
import treePlotter
import sklearn.datasets as datasets
from sklearn.metrics import mean_squared_error
import sklearn.tree as tree
import graphvizclass DecisionTreeRegressor:def __init__(self, min_samples_split=3,min_samples_leaf=1,random_state=False):self.min_samples_split=min_samples_splitself.min_samples_leaf=min_samples_leafself.random=random_stateself.tree = Nonedef fit(self, X, y):D = {}D['X'] = XD['y'] = yA = np.arange(X.shape[1])self.tree = self.TreeGenerate(D, A)def predict(self, X):if self.tree is None:raise RuntimeError('cant predict before fit')y_pred = []for i in range(X.shape[0]):tree = self.treex = X[i]while True:if not isinstance(tree, dict):y_pred.append(tree)breaka = list(tree.keys())[0]tree = tree[a]if isinstance(tree, dict):val = x[a]split_val=float(list(tree.keys())[0][1:])if val<=split_val:tree=tree[list(tree.keys())[0]]else:tree=tree[list(tree.keys())[1]]else:y_pred.append(tree)breakreturn np.array(y_pred)def TreeGenerate(self, D, A):X = D['X']y = D['y']if len(y)<=self.min_samples_split:return np.mean(y)split_j=Nonesplit_s=Nonemin_val=1.e10select_A=Aif self.random is True:d=len(A)select_A=np.random.choice(A,size=int(d//2),replace=False)for j in select_A:for s in np.unique(X[:,j]):left_indices=np.where(X[:,j]<=s)[0]right_indices=np.where(X[:,j]>s)[0]if len(left_indices)<self.min_samples_leaf or len(right_indices)<self.min_samples_leaf:continueval=np.sum((y[left_indices]-np.mean(y[left_indices]))**2)+np.sum((y[right_indices]-np.mean(y[right_indices]))**2)if val<min_val:split_j=jsplit_s=smin_val=valif split_j is None:return np.mean(y)tree = {split_j: {}}left_indices=np.where(X[:,split_j]<=split_s)[0]right_indices=np.where(X[:,split_j]>split_s)[0]D_left, D_right = {},{}D_left['X'],D_left['y'] = X[left_indices],y[left_indices]D_right['X'],D_right['y']=X[right_indices],y[right_indices]tree[split_j]['l'+str(split_s)]=self.TreeGenerate(D_left,A)tree[split_j]['r'+str(split_s)]=self.TreeGenerate(D_right,A)# 当前节点值tree[split_j]['val']=np.mean(y)return treeif __name__=='__main__':breast_data = datasets.load_boston()X, y = breast_data.data, breast_data.targetX_train, y_train = X[:200], y[:200]X_test, y_test = X[200:], y[200:]decisiontree_reg=DecisionTreeRegressor(min_samples_split=20,min_samples_leaf=5)decisiontree_reg.fit(X_train,y_train)print(decisiontree_reg.tree)treePlotter.createPlot(decisiontree_reg.tree)y_pred=decisiontree_reg.predict(X_test)print('tinyml mse:',mean_squared_error(y_test,y_pred))sklearn_reg=tree.DecisionTreeRegressor(min_samples_split=20,min_samples_leaf=5,random_state=False)sklearn_reg.fit(X_train,y_train)print(sklearn_reg.feature_importances_)sklearn_pred=sklearn_reg.predict(X_test)print('sklearn mse:',mean_squared_error(y_test,sklearn_pred))dot_data=tree.export_graphviz(sklearn_reg,out_file=None)graph=graphviz.Source(dot_data)4.9