一、分子图
分子图(molecular graph)是一种用来表示分子结构的图形方式,其中原子被表示为节点(vertices),化学键被表示为边(edges)。对于HIV(人类免疫缺陷病毒),分子图可以用来详细描述其复杂的化学结构和相互作用,这对于理解HIV的生物学特性和开发治疗药物至关重要。
二、分子图分类
直接撸代码,以HIV为例
HIV分子图一共有8个问题,现在取第一个问题:
"The human immunodeficiency viruses (HIV) are a type of retrovirus, which induce acquired immune deficiency syndrome (AIDs). Now there are six main classes of antiretroviral drugs to treating AIDs patients approved by FDA, which are the nucleoside reverse transcriptase inhibitors (NRTIs), the non-nucleoside reverse transcriptase inhibitors (NNRTIs), the protease inhibitors, the integrase inhibitor, the fusion inhibitor, and the chemokine receptor CCR5 antagonist. Due to the missing 3\u2019hydroxyl group, NRTIs prevent the formation of a 3\u2019-5\u2019-phosphodiester bond in growing DNA chains. The hydroxyl group of the inhibitor interacts with the carboxyl group of the protease active site residues, Asp 25 and Asp 25\u2032, by hydrogen bonds. The inhibitor-contacting residues of HIV protease are relatively conserved, including Gly 27, Asp 29, Asp 30, and Gly 48. Is this molecule effective to this assay?",
Answer is yes or no.
针对上面问题,进行二分类
代码环节
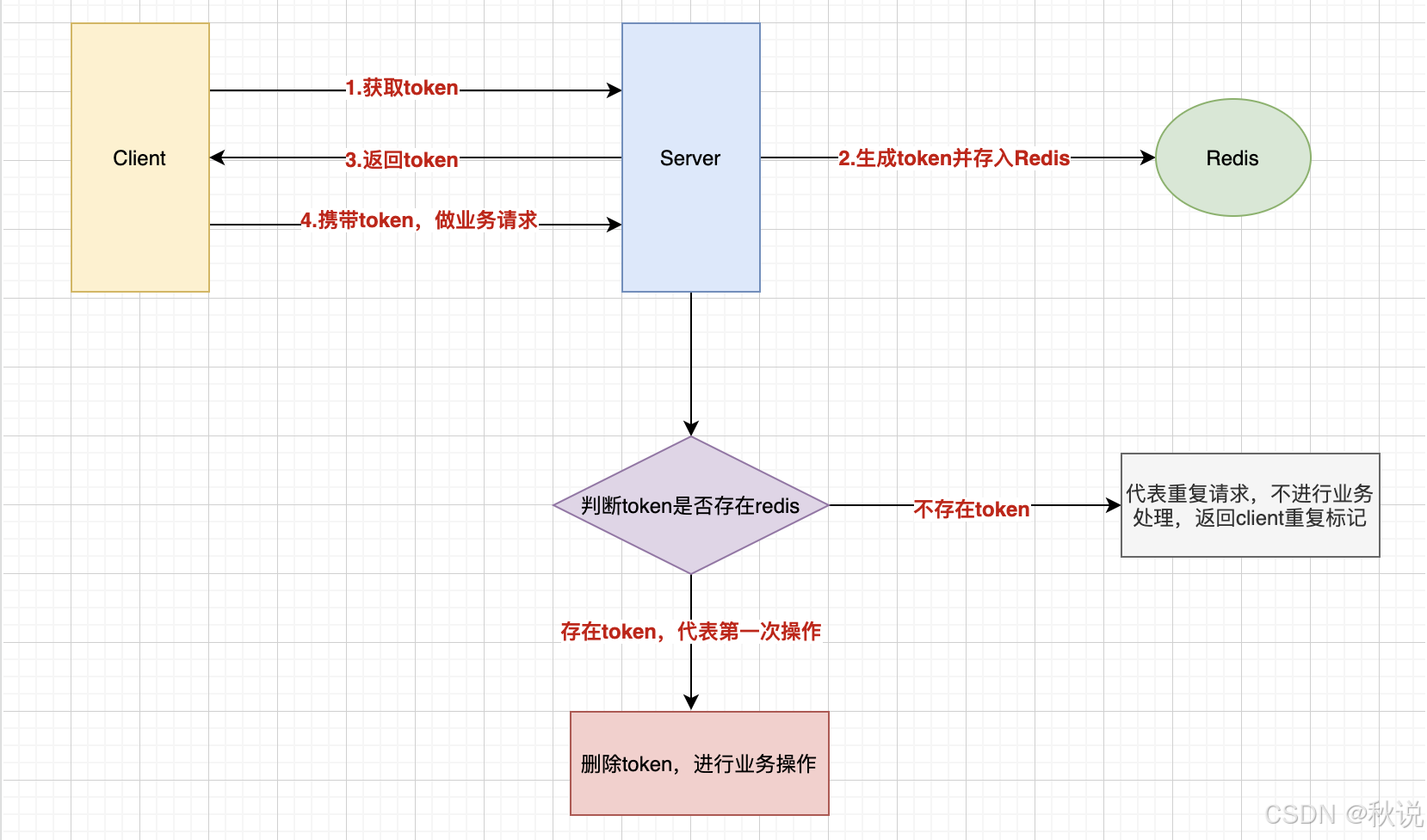
oversample_data.py 数据正负样本均衡处理,进行上采样
在机器学习和统计学中,上采样(Upsampling)和下采样(Downsampling)是两种常用的数据预处理技术,它们用于处理数据集中的类别不平衡问题。类别不平衡指的是数据集中某些类别的样本数量远多于其他类别,这可能会导致模型偏向于多数类别,从而影响模型的泛化能力和公平性。以下是上采样和下采样的基本概念:
上采样(Upsampling)
上采样是指增加少数类别(underrepresented classes)的样本数量,使其与多数类别(overrepresented classes)的样本数量相匹配或接近。这样做的目的是减少模型对多数类别的偏好,提高对少数类别的识别能力。上采样可以通过以下几种方式实现:
- 简单复制:直接复制少数类别的样本,直到其数量与多数类别相等。
- 随机采样:从少数类别中随机抽取样本,直到达到所需的数量。
- SMOTE(Synthetic Minority Over-sampling Technique):这是一种更复杂的技术,它通过在少数类别的样本之间插入新的、合成的样本来增加样本数量。这些合成样本是通过在少数类别样本的k-最近邻之间进行插值来生成的。
下采样(Downsampling)
下采样是指减少多数类别的样本数量,使其与少数类别的样本数量相匹配或接近。这种方法的目的是减少训练集中的样本数量,以避免模型过度拟合到多数类别。下采样可以通过以下几种方式实现:
- 简单随机采样:从多数类别中随机选择一定数量的样本,丢弃其余的样本。
- 聚类中心采样:使用聚类算法找到多数类别的中心点,然后只保留这些中心点附近的样本。
- ** Tomek Links**:这是一种特殊的下采样技术,它识别并删除那些与少数类别样本非常接近的多数类别样本,因为这些样本可能会导致模型学习到错误的边界。
应用场景
- 上采样:当数据集中存在严重的类别不平衡,且我们希望模型能够对少数类别有较好的识别能力时,上采样是一个合适的选择。
- 下采样:当数据集中的类别不平衡不是特别严重,或者我们更关注模型的整体性能而不是对少数类别的识别能力时,下采样可能是一个更好的选择。
注意事项
- 上采样和下采样都有其局限性。上采样可能会导致过拟合,因为它增加了数据集中的样本数量,尤其是合成样本。下采样可能会导致信息丢失,因为它丢弃了一部分数据。
- 在实际应用中,可能需要结合上采样和下采样,或者使用其他技术(如调整类别权重、集成学习等)来处理类别不平衡问题。
import pandas as pddata = pd.read_csv("data/raw/HIV_train.csv")
data.index = data["index"]
data["HIV_active"].value_counts()
start_index = data.iloc[0]["index"]# %% Apply oversampling# Check how many additional samples we need
neg_class = data["HIV_active"].value_counts()[0]
pos_class = data["HIV_active"].value_counts()[1]
multiplier = int(neg_class/pos_class) - 1# Replicate the dataset for the positive class
replicated_pos = [data[data["HIV_active"] == 1]]*multiplier# Append replicated data
data = data.append(replicated_pos,ignore_index=True)
print(data.shape)# Shuffle dataset
data = data.sample(frac=1).reset_index(drop=True)# Re-assign index (This is our ID later)
index = range(start_index, start_index + data.shape[0])
data.index = index
data["index"] = data.index
data.head()# Save
data.to_csv("data/raw/HIV_train_oversampled.csv")
特征转化 dataset_featurizer.py ,生成pyg格式的Data对象
构图主要包含:
node_feature,从9个维度描述分子结构的信息
edge_feature,描述边的信息
edge_index
label
import pandas as pd
import torch
import torch_geometric
from torch_geometric.data import Dataset, Data
import os
from tqdm import tqdm
import deepchem as dc
from rdkit import Chemprint(f"Torch version: {torch.__version__}")
print(f"Cuda available: {torch.cuda.is_available()}")
print(f"Torch geometric version: {torch_geometric.__version__}")class MoleculeDataset(Dataset):def __init__(self, root, filename, test=False, transform=None, pre_transform=None):"""root = Where the dataset should be stored. This folder is splitinto raw_dir (downloaded dataset) and processed_dir (processed data). """self.test = testself.filename = filenamesuper(MoleculeDataset, self).__init__(root, transform, pre_transform)@propertydef raw_file_names(self):""" If this file exists in raw_dir, the download is not triggered.(The download func. is not implemented here) """return self.filename@propertydef processed_file_names(self):""" If these files are found in raw_dir, processing is skipped"""self.data = pd.read_csv(os.path.join(self.root,self.filename)).reset_index()if self.test:return [f'data_test_{i}.pt' for i in list(self.data.index)]else:return [f'data_{i}.pt' for i in list(self.data.index)]def download(self):passdef process(self):self.data = pd.read_csv(os.path.join(self.root,self.filename))for index, mol in tqdm(self.data.iterrows(), total=self.data.shape[0]):mol_obj = Chem.MolFromSmiles(mol["smiles"])# Get node featuresnode_feats = self._get_node_features(mol_obj)# Get edge featuresedge_feats = self._get_edge_features(mol_obj)# Get adjacency infoedge_index = self._get_adjacency_info(mol_obj)# Get labels infolabel = self._get_labels(mol["HIV_active"])# Create data objectdata = Data(x=node_feats, edge_index=edge_index,edge_attr=edge_feats,y=label,smiles=mol["smiles"]) if self.test:torch.save(data, os.path.join(self.processed_dir, f'data_test_{index}.pt'))else:torch.save(data, os.path.join(self.processed_dir, f'data_{index}.pt'))def _get_node_features(self, mol):""" This will return a matrix / 2d array of the shape[Number of Nodes, Node Feature size]"""all_node_feats = []for atom in mol.GetAtoms():node_feats = []# Feature 1: Atomic number node_feats.append(atom.GetAtomicNum())# Feature 2: Atom degreenode_feats.append(atom.GetDegree())# Feature 3: Formal chargenode_feats.append(atom.GetFormalCharge())# Feature 4: Hybridizationnode_feats.append(atom.GetHybridization())# Feature 5: Aromaticitynode_feats.append(atom.GetIsAromatic())# Feature 6: Total Num Hsnode_feats.append(atom.GetTotalNumHs())# Feature 7: Radical Electronsnode_feats.append(atom.GetNumRadicalElectrons())# Feature 8: In Ringnode_feats.append(atom.IsInRing())# Feature 9: Chiralitynode_feats.append(atom.GetChiralTag())# Append node features to matrixall_node_feats.append(node_feats)all_node_feats = np.asarray(all_node_feats)return torch.tensor(all_node_feats, dtype=torch.float)def _get_edge_features(self, mol):""" This will return a matrix / 2d array of the shape[Number of edges, Edge Feature size]"""all_edge_feats = []for bond in mol.GetBonds():edge_feats = []# Feature 1: Bond type (as double)edge_feats.append(bond.GetBondTypeAsDouble())# Feature 2: Ringsedge_feats.append(bond.IsInRing())# Append node features to matrix (twice, per direction)all_edge_feats += [edge_feats, edge_feats]all_edge_feats = np.asarray(all_edge_feats)return torch.tensor(all_edge_feats, dtype=torch.float)def _get_adjacency_info(self, mol):"""We could also use rdmolops.GetAdjacencyMatrix(mol)but we want to be sure that the order of the indicesmatches the order of the edge features"""edge_indices = []for bond in mol.GetBonds():i = bond.GetBeginAtomIdx()j = bond.GetEndAtomIdx()edge_indices += [[i, j], [j, i]]edge_indices = torch.tensor(edge_indices)edge_indices = edge_indices.t().to(torch.long).view(2, -1)return edge_indicesdef _get_labels(self, label):label = np.asarray([label])return torch.tensor(label, dtype=torch.int64)def len(self):return self.data.shape[0]def get(self, idx):""" - Equivalent to __getitem__ in pytorch- Is not needed for PyG's InMemoryDataset"""if self.test:data = torch.load(os.path.join('../',self.processed_dir, f'data_test_{idx}.pt'))else:data = torch.load(os.path.join('../', self.processed_dir, f'data_{idx}.pt')) return data模型结构 :GNN1,常见的GAT结构
import os
import rdkit
import torch
import cairosvg
import numpy as np
import torch_geometric
import pandas as pd
from tqdm import tqdm
import deepchem as dc
from PIL import Image
from rdkit import Chem
from rdkit import Chem
from rdkit import RDConfig
from rdkit.Chem import Draw
from rdkit.Chem import Draw
from rdkit.Chem import rdBase
import matplotlib.pyplot as plt
import IPython.display as display
from rdkit.Chem.Draw import IPythonConsole
from sklearn.model_selection import train_test_split
from torch_geometric.data import Dataset, Data
from torch_geometric.data import DataLoader
from dataset_featurizer import MoleculeDataset
import torch.nn.functional as F
import seaborn as sns
from torch.nn import Sequential, Linear, BatchNorm1d, ModuleList, ReLU
from torch_geometric.nn import TransformerConv, TopKPooling, GATConv, BatchNorm
from torch_geometric.nn import global_mean_pool as gap, global_max_pool as gmp
from sklearn.metrics import confusion_matrix, f1_score, \accuracy_score, precision_score, recall_score, roc_auc_score
from torch_geometric.nn.conv.x_conv import XConv
torch.manual_seed(42)class GNN1(torch.nn.Module):def __init__(self, feature_size):super(GNN1, self).__init__()num_classes =2embedding_size = 1024# GNN1 layersself.conv1 = GATConv(feature_size, embedding_size, heads = 3, dropout = 0.3)self.head_transform1 = Linear(embedding_size*3, embedding_size)self.pool1 = TopKPooling(embedding_size, ratio=0.8)self.conv2 = GATConv(feature_size, embedding_size, heads = 3, dropout = 0.3)self.head_transform2 = Linear(embedding_size*3, embedding_size)self.pool2 = TopKPooling(embedding_size, ratio=0.5)self.conv3 = GATConv(feature_size, embedding_size, heads = 3, dropout = 0.3)self.head_transform3 = Linear(embedding_size*3, embedding_size)self.pool3 = TopKPooling(embedding_size, ratio=0.3)# Linear layersself.linear1 = Linear(embedding_size*2, 1024)self.linear2 = Linear(1024, num_classes)def forward(self, x, edge_attr, edge_index, batch_index):# First blockx = self.conv1(x,edge_index)x = self.head_transform1(x)x, edge_index, edge_attr, batch_index, _,_ = self.pool1(x,edge_index,None,batch_index)x1 = torch.cat([gap(x,batch_index), gap(x,batch_index)], dim=1)# Second blockx = self.conv2(x,edge_index)x = self.head_transform2(x)x, edge_index, edge_attr, batch_index, _,_ = self.pool2(x,edge_index,None,batch_index)x2 = torch.cat([gap(x,batch_index), gap(x,batch_index)], dim=1)# Third blockx = self.conv3(x,edge_index)x = self.head_transform3(x)x, edge_index, edge_attr, batch_index, _,_ = self.pool3(x,edge_index,None,batch_index)x3 = torch.cat([gap(x,batch_index), gap(x,batch_index)], dim=1)# Concat pooled vectorx = x1+x2+x3# Output blockx = self.linear1(x).relu()x = F.dropout(x,p=0.5,training=self.training)x = self.linear2(x)return xGNN2
import os
import rdkit
import torch
import cairosvg
import numpy as np
import torch_geometric
import pandas as pd
from tqdm import tqdm
import deepchem as dc
from PIL import Image
from rdkit import Chem
from rdkit import Chem
from rdkit import RDConfig
from rdkit.Chem import Draw
from rdkit.Chem import Draw
from rdkit.Chem import rdBase
import matplotlib.pyplot as plt
import IPython.display as display
from rdkit.Chem.Draw import IPythonConsole
from sklearn.model_selection import train_test_split
from torch_geometric.data import Dataset, Data
from torch_geometric.data import DataLoader
from dataset_featurizer import MoleculeDataset
import torch.nn.functional as F
import seaborn as sns
from torch.nn import Sequential, Linear, BatchNorm1d, ModuleList, ReLU
from torch_geometric.nn import TransformerConv, TopKPooling, GATConv, BatchNorm
from torch_geometric.nn import global_mean_pool as gap, global_max_pool as gmp
from sklearn.metrics import confusion_matrix, f1_score, \accuracy_score, precision_score, recall_score, roc_auc_score
from torch_geometric.nn.conv.x_conv import XConv
torch.manual_seed(42)class GNN2(torch.nn.Module):def __init__(self, feature_size):super(GNN2, self).__init__()num_classes = 2embedding_size = 1024# GNN2 layersself.conv1 = GATConv(feature_size, embedding_size, heads=3, dropout=0.3)self.head_transform1 = Linear(embedding_size*3, embedding_size)self.pool1 = TopKPooling(embedding_size, ratio=0.8)self.conv2 = GATConv(embedding_size, embedding_size, heads=3, dropout=0.3)self.head_transform2 = Linear(embedding_size*3, embedding_size)self.pool2 = TopKPooling(embedding_size, ratio=0.5)self.conv3 = GATConv(embedding_size, embedding_size, heads=3, dropout=0.3)self.head_transform3 = Linear(embedding_size*3, embedding_size)self.pool3 = TopKPooling(embedding_size, ratio=0.3)# Transformer layerself.transformer = TransformerConv(embedding_size, heads=4, num_layers=3)# Isomorphism layerself.isomorphism = XConv(Linear(feature_size, embedding_size), K=3)# Linear layersself.linear1 = Linear(embedding_size*2, 1024)self.linear2 = Linear(1024, num_classes)def forward(self, x, edge_attr, edge_index, batch_index):# Isomorphism layerx = self.isomorphism(x, edge_index, edge_attr)# Transformer layerx = self.transformer(x, edge_index)# First blockx = self.conv1(x, edge_index)x = self.head_transform1(x)x, edge_index, edge_attr, batch_index, _, _ = self.pool1(x,edge_index,None,batch_index)x1 = torch.cat([gap(x, batch_index), gap(x, batch_index)], dim=1)# Second blockx = self.conv2(x, edge_index)x = self.head_transform2(x)x, edge_index, edge_attr, batch_index, _, _ = self.pool2(x,edge_index,None,batch_index)x2 = torch.cat([gap(x, batch_index), gap(x, batch_index)], dim=1)# Third blockx = self.conv3(x, edge_index)x = self.head_transform3(x)x, edge_index, edge_attr, batch_index, _, _ = self.pool3(x,edge_index,None,batch_index)x3 = torch.cat([gap(x, batch_index), gap(x, batch_index)], dim=1)# Concat pooled vectorx = x1 + x2 + x3# Output blockx = self.linear1(x).relu()x = F.dropout(x, p=0.5, training=self.training)x = self.linear2(x)return xGNN3
import os
import rdkit
import torch
import cairosvg
import numpy as np
import torch_geometric
import pandas as pd
from tqdm import tqdm
import deepchem as dc
from PIL import Image
from rdkit import Chem
from rdkit import Chem
from rdkit import RDConfig
from rdkit.Chem import Draw
from rdkit.Chem import Draw
from rdkit.Chem import rdBase
import matplotlib.pyplot as plt
import IPython.display as display
from rdkit.Chem.Draw import IPythonConsole
from sklearn.model_selection import train_test_split
from torch_geometric.data import Dataset, Data
from torch_geometric.data import DataLoader
from dataset_featurizer import MoleculeDataset
import torch.nn.functional as F
import seaborn as sns
from torch.nn import Sequential, Linear, BatchNorm1d, ModuleList, ReLU
from torch_geometric.nn import TransformerConv, TopKPooling, GATConv, BatchNorm
from torch_geometric.nn import global_mean_pool as gap, global_max_pool as gmp
from sklearn.metrics import confusion_matrix, f1_score, \accuracy_score, precision_score, recall_score, roc_auc_score
from torch_geometric.nn.conv.x_conv import XConv
torch.manual_seed(42)class GNN3(torch.nn.Module):def __init__(self, feature_size, edge_feature_size):super(GNN3, self).__init__()num_classes = 2embedding_size = 1024# GNN3 layersself.conv1 = GATConv(feature_size, embedding_size, heads=3, dropout=0.3)self.head_transform1 = Linear(embedding_size*3 + edge_feature_size, embedding_size)self.pool1 = TopKPooling(embedding_size, ratio=0.8)self.conv2 = GATConv(embedding_size, embedding_size, heads=3, dropout=0.3)self.head_transform2 = Linear(embedding_size*3 + edge_feature_size, embedding_size)self.pool2 = TopKPooling(embedding_size, ratio=0.5)self.conv3 = GATConv(embedding_size, embedding_size, heads=3, dropout=0.3)self.head_transform3 = Linear(embedding_size*3 + edge_feature_size, embedding_size)self.pool3 = TopKPooling(embedding_size, ratio=0.3)# Transformer layerself.transformer = TransformerConv(embedding_size, heads=4, num_layers=3)# Isomorphism layerself.isomorphism = XConv(Linear(feature_size + edge_feature_size, embedding_size), K=3)# Linear layersself.linear1 = Linear(embedding_size*2, 1024)self.linear2 = Linear(1024, num_classes)def forward(self, x, edge_attr, edge_index, batch_index):# Isomorphism layerx = self.isomorphism(x, edge_index, edge_attr)# Transformer layerx = self.transformer(x, edge_index)# First blockx_with_edge = torch.cat([x, edge_attr], dim=1)x = self.conv1(x_with_edge, edge_index)x = self.head_transform1(x)x, edge_index, edge_attr, batch_index, _, _ = self.pool1(x,edge_index,edge_attr,batch_index)x1 = torch.cat([gap(x, batch_index), gap(x, batch_index)], dim=1)# Second blockx_with_edge = torch.cat([x, edge_attr], dim=1)x = self.conv2(x_with_edge, edge_index)x = self.head_transform2(x)x, edge_index, edge_attr, batch_index, _, _ = self.pool2(x,edge_index,edge_attr,batch_index)x2 = torch.cat([gap(x, batch_index), gap(x, batch_index)], dim=1)# Third blockx_with_edge = torch.cat([x, edge_attr], dim=1)x = self.conv3(x_with_edge, edge_index)x = self.head_transform3(x)x, edge_index, edge_attr, batch_index, _, _ = self.pool3(x,edge_index,edge_attr,batch_index)x3 = torch.cat([gap(x, batch_index), gap(x, batch_index)], dim=1)# Concat pooled vectorx = x1 + x2 + x3# Output blockx = self.linear1(x).relu()x = F.dropout(x, p=0.5, training=self.training)x = self.linear2(x)return xtrain.py 训练流程代码
#%%
import os
import rdkit
import torch
import cairosvg
import numpy as np
import torch_geometric
import pandas as pd
from tqdm import tqdm
import deepchem as dc
from PIL import Image
from rdkit import Chem
from rdkit import Chem
from rdkit import RDConfig
from rdkit.Chem import Draw
from rdkit.Chem import Draw
from rdkit.Chem import rdBase
import matplotlib.pyplot as plt
import IPython.display as display
from rdkit.Chem.Draw import IPythonConsole
from sklearn.model_selection import train_test_split
from torch_geometric.data import Dataset, Data
from torch_geometric.data import DataLoader
from dataset_featurizer import MoleculeDataset
from gnn_project.model.GNN1 import GNN1
from gnn_project.model.GNN2 import GNN2
from gnn_project.model.GNN3 import GNN3
import torch.nn.functional as F
import seaborn as sns
from torch.nn import Sequential, Linear, BatchNorm1d, ModuleList, ReLU
from torch_geometric.nn import TransformerConv, TopKPooling, GATConv, BatchNorm
from torch_geometric.nn import global_mean_pool as gap, global_max_pool as gmp
from sklearn.metrics import confusion_matrix, f1_score, \accuracy_score, precision_score, recall_score, roc_auc_score
from torch_geometric.nn.conv.x_conv import XConv
torch.manual_seed(42)data_path = 'data/raw/HIV_data.csv'
data = pd.read_csv(data_path,index_col=[0])
print("###### Raw Data Shape - ")
print(data.shape)
print("0 : ",data["HIV_active"].value_counts()[0])
print("1 : ",data["HIV_active"].value_counts()[1])#%%
# Split the data, Due to imbalance datasets we need to set train ration high
print("###### After Data split Shape - ")
train_data = pd.read_csv('data/split_data/HIV_train.csv')
test_data = pd.read_csv('data/split_data/HIV_test.csv')
train_data_oversampled = pd.read_csv('data/split_data/HIV_train_oversampled.csv')
print("Train Data ", train_data.shape)
print("0 : ",train_data["HIV_active"].value_counts()[0])
print("1 : ",train_data["HIV_active"].value_counts()[1])print("Test Data ", test_data.shape)
print("0 : ",test_data["HIV_active"].value_counts()[0])
print("1 : ",test_data["HIV_active"].value_counts()[1])print("Train Data Oversampled ", train_data_oversampled.shape)
print("0 : ",train_data_oversampled["HIV_active"].value_counts()[0])
print("1 : ",train_data_oversampled["HIV_active"].value_counts()[1])# %%
# Define the folder path to save the images
output_folder = "visualization"
os.makedirs(output_folder, exist_ok=True)sample_smiles = train_data["smiles"][4:30].values
sdf = Chem.SDMolSupplier(output_folder+'/cdk2.sdf')
mols = [m for m in sdf]for i, smiles in enumerate(sample_smiles):core = Chem.MolFromSmiles(smiles)img = Draw.MolsToGridImage(mols, molsPerRow=3, highlightAtomLists=[mol.GetSubstructMatch(core) for mol in mols], useSVG=True)## Save the image in the output folderimage_path = os.path.join(output_folder, f"image_{i}.png")cairosvg.svg2png(bytestring=img.data, write_to=image_path)breakprint(f"Image saved: {image_path}")#%%
print("######## Loading dataset...")
train_dataset = MoleculeDataset(root="data/split_data", filename="HIV_train_oversampled.csv")
test_dataset = MoleculeDataset(root="data/split_data", filename="HIV_test.csv", test=True) # %%
print("######## Loading GNN1 Model...")
device = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
def count_parameters(model):return sum(p.numel() for p in model.parameters() if p.requires_grad)model = GNN1(feature_size=train_dataset[0].x.shape[1])
model = model.to(device)
print(f"######### Number of parameters: {count_parameters(model)}")
print(model)
# %%
# Loss and Optimizer
# Due to imbalance postive and negative label so apply weight in the +ve side < 1 increases precision, > 1 recall
weight = torch.tensor([1,10], dtype=torch.float32).to(device)
loss_fn = torch.nn.CrossEntropyLoss(weight==weight)
optimizer = torch.optim.SGD(model.parameters(), lr=0.1,momentum=0.9)
scheduler = torch.optim.lr_scheduler.ExponentialLR(optimizer, gamma=0.95)# %%
NUM_GRAPHS_PER_BATCH = 256
train_loader = DataLoader(train_dataset,batch_size = NUM_GRAPHS_PER_BATCH, shuffle=True)
test_loader = DataLoader(test_dataset,batch_size = NUM_GRAPHS_PER_BATCH, shuffle=True)#%%#%%def train(epoch, model, train_loader, optimizer, loss_fn):#Enumerate over the dataall_preds = []all_labels = []running_loss = 0.0step = 0for _,batch in enumerate(tqdm(train_loader)):#Using GPUbatch.to(device)#Reset gradientoptimizer.zero_grad()#passing the node feature and concat the infopred = model(batch.x.float(),batch.edge_attr.float,batch.edge_index,batch.batch)# Calculating the loss and gradientsloss = loss_fn(torch.squeeze(pred), batch.y.float())loss.backward() optimizer.step() # Update trackingrunning_loss += loss.item()step += 1all_preds.append(np.rint(torch.sigmoid(pred).cpu().detach().numpy()))all_labels.append(batch.y.cpu().detach().numpy())all_preds = np.concatenate(all_preds).ravel()all_labels = np.concatenate(all_labels).ravel()calculate_metrics(all_preds, all_labels, epoch, "train")return running_loss/step
#%%
def test(epoch, model, test_loader, loss_fn):#Enumerate over the dataall_preds = []all_preds_raw = []all_labels = []running_loss = 0.0step = 0for batch in test_loader:#Using GPUbatch.to(device)pred = model(batch.x.float(),batch.edge_attr.float,batch.edge_index,batch.batch)loss = loss_fn(torch.squeeze(pred), batch.y.float()) # Update trackingrunning_loss += loss.item()step += 1all_preds.append(np.rint(torch.sigmoid(pred).cpu().detach().numpy()))all_preds_raw.append(torch.sigmoid(pred).cpu().detach().numpy())all_labels.append(batch.y.cpu().detach().numpy())all_preds = np.concatenate(all_preds).ravel()all_labels = np.concatenate(all_labels).ravel()print(all_preds_raw[0][:10])print(all_preds[:10])print(all_labels[:10])calculate_metrics(all_preds, all_labels, epoch, "test")log_conf_matrix(all_preds, all_labels, epoch)return running_loss/step#%%def log_conf_matrix(y_pred, y_true, epoch):# Log confusion matrix as imagecm = confusion_matrix(y_pred, y_true)classes = ["0", "1"]df_cfm = pd.DataFrame(cm, index = classes, columns = classes)plt.figure(figsize = (10,7))cfm_plot = sns.heatmap(df_cfm, annot=True, cmap='Blues', fmt='g')cfm_plot.figure.savefig(f'data/images/cm_{epoch}.png')def calculate_metrics(y_pred, y_true, epoch, type):print(f"\n Confusion matrix: \n {confusion_matrix(y_pred, y_true)}")print(f"F1 Score: {f1_score(y_true, y_pred)}")print(f"Accuracy: {accuracy_score(y_true, y_pred)}")prec = precision_score(y_true, y_pred)rec = recall_score(y_true, y_pred)print(f"Precision: {prec}")print(f"Recall: {rec}")try:roc = roc_auc_score(y_pred,y_true)print(f"ROC AUC: {roc}")except:print(f"ROC AUC: not definded")
# %%
# Start training
print("###### Start GNN Model training")
best_loss = 1000
early_stopping_counter = 0
for epoch in range(300): if early_stopping_counter <= 10: # = x * 5 # Trainingmodel.train()loss = train(epoch, model, train_loader, optimizer, loss_fn)print(f"Epoch {epoch} | Train Loss {loss}")#mlflow.log_metric(key="Train loss", value=float(loss), step=epoch)# Testingmodel.eval()if epoch % 5 == 0:loss = test(epoch, model, test_loader, loss_fn)print(f"Epoch {epoch} | Test Loss {loss}")# mlflow.log_metric(key="Test loss", value=float(loss), step=epoch)# Update best lossif float(loss) < best_loss:best_loss = loss# Save the currently best model #mlflow.pytorch.log_model(model, "model", signature=SIGNATURE)early_stopping_counter = 0else:early_stopping_counter += 1scheduler.step()else:print("Early stopping due to no improvement.")print([best_loss])
print(f"Finishing training with best test loss: {best_loss}")
print([best_loss])output_folder = "model_weight"
os.makedirs(output_folder, exist_ok=True)
model_path = os.join.path(output_folder,"model.pth") # Replace with the desired path to save the model
torch.save(model, model_path)# %%train_optimization.py
import argparse
import os
import pandas as pd
import torch
from sklearn.metrics import confusion_matrix, f1_score, accuracy_score, precision_score, recall_score, roc_auc_score
from torch_geometric.data import DataLoader
from torch.nn import BCELoss
from torch.optim import Adam
from dataset_featurizer import MoleculeDataset
from model.GNN1 import GNN1
from model.GNN2 import GNN2
from model.GNN3 import GNN3
import optunatorch.manual_seed(42)# Create a parser object
parser = argparse.ArgumentParser(description='GNN Model Training')# Add arguments for data paths
parser.add_argument('--test_data_path', type=str, required=True, help='Path to the test data file')
parser.add_argument('--train_oversampled', type=str, required=True, help='Path to the train oversampled data file')# Add an argument for the GNN model selection
parser.add_argument('--model', type=str, choices=['GNN1', 'GNN2', 'GNN3'], default='GNN1', help='Choose the GNN model (GNN1, GNN2, GNN3)')# Add an argument for the number of epochs
parser.add_argument('--epochs', type=int, default=100, help='Number of training epochs')# Parse the command-line arguments
args = parser.parse_args()# Get the data paths from the command-line arguments
test_data_path = args.test_data_path
train_oversampled_ = args.train_oversampled# Get the selected model from the command-line arguments
selected_model = args.model# Get the number of epochs from the command-line arguments
num_epochs = args.epochs# Load the data
test_data = pd.read_csv(test_data_path)
train_data = pd.read_csv(train_oversampled)model_folder = "model_weights"
os.makedirs(model_folder, exist_ok=True)# Define the GNN model based on the selected model
if selected_model == 'GNN1':model = GNN1(feature_size=train_data[0].x.shape[1])elif selected_model == 'GNN2':model = GNN2(feature_size=train_data[0].x.shape[1])elif selected_model == 'GNN3':model = GNN3(feature_size=train_data[0].x.shape[1])else:raise ValueError('Invalid model selected')# Define the loss function
loss_fn = BCELoss()# Define the optimizer
optimizer = Adam(model.parameters(), lr=0.001)# Define the objective function for Optuna optimization
def objective(trial):# Sample the hyperparameters to be tunedhyperparameters = {"batch_size": trial.suggest_categorical("batch_size", [32, 128, 64]),"learning_rate": trial.suggest_loguniform("learning_rate", 1e-4, 1e-1),"weight_decay": trial.suggest_loguniform("weight_decay", 1e-5, 1e-3),"sgd_momentum": trial.suggest_uniform("sgd_momentum", 0.5, 0.9),"scheduler_gamma": trial.suggest_categorical("scheduler_gamma", [0.995, 0.9, 0.8, 0.5, 1]),"pos_weight": trial.suggest_categorical("pos_weight", [1.0]),"model_embedding_size": trial.suggest_categorical("model_embedding_size", [8, 16, 32, 64, 128]),"model_attention_heads": trial.suggest_int("model_attention_heads", 1, 4),"model_layers": trial.suggest_categorical("model_layers", [3]),"model_dropout_rate": trial.suggest_uniform("model_dropout_rate", 0.2, 0.9),"model_top_k_ratio": trial.suggest_categorical("model_top_k_ratio", [0.2, 0.5, 0.8, 0.9]),"model_top_k_every_n": trial.suggest_categorical("model_top_k_every_n", [0]),"model_dense_neurons": trial.suggest_categorical("model_dense_neurons", [16, 128, 64, 256, 32]),}# Set the hyperparameters in the modelmodel.set_hyperparameters(**hyperparameters)# Create the data loaderstrain_loader = DataLoader(train_dataset, batch_size=hyperparameters["batch_size"], shuffle=True)test_loader = DataLoader(test_dataset, batch_size=hyperparameters["batch_size"], shuffle=False)# Train the modelfor epoch in range(num_epochs):model.train()for batch in train_loader:optimizer.zero_grad()out = model(batch)loss = loss_fn(out, batch.y)loss.backward()optimizer.step()# Evaluate the modelmodel.eval()y_true = []y_pred = []with torch.no_grad():for batch in test_loader:out = model(batch)pred = (out >= 0.5).float()y_pred.extend(pred.tolist())y_true.extend(batch.y.tolist())# Compute evaluation metricsaccuracy = accuracy_score(y_true, y_pred)precision = precision_score(y_true, y_pred)recall = recall_score(y_true, y_pred)f1 = f1_score(y_true, y_pred)auc_roc = roc_auc_score(y_true, y_pred)return f1# Create the dataset
test_dataset = MoleculeDataset(test_data)
train_dataset = MoleculeDataset(train_data)# Create the Optuna study and run the optimization
study = optuna.create_study(direction='maximize')
study.optimize(objective, n_trials=100)# Get the best hyperparameters and metric
best_hyperparameters = study.best_params
best_metric = study.best_value# Set the best hyperparameters in the model
model.set_hyperparameters(**best_hyperparameters)# Create the best data loaders
best_train_loader = DataLoader(train_dataset, batch_size=best_hyperparameters["batch_size"], shuffle=True)
best_test_loader = DataLoader(test_dataset, batch_size=best_hyperparameters["batch_size"], shuffle=False)# Train the model using the best hyperparameters
for epoch in range(num_epochs):model.train()for batch in best_train_loader:optimizer.zero_grad()out = model(batch)loss = loss_fn(out, batch.y)loss.backward()optimizer.step()# Evaluate the modelmodel.eval()y_true = []y_pred = []with torch.no_grad():for batch in best_test_loader:out = model(batch)pred = (out >= 0.5).float()y_pred.extend(pred.tolist())y_true.extend(batch.y.tolist())# Compute evaluation metricsaccuracy = accuracy_score(y_true, y_pred)precision = precision_score(y_true, y_pred)recall = recall_score(y_true, y_pred)f1 = f1_score(y_true, y_pred)auc_roc = roc_auc_score(y_true, y_pred)print(f'Epoch {epoch + 1}: Accuracy={accuracy:.4f}, Precision={precision:.4f}, Recall={recall:.4f}, F1={f1:.4f}, AUC-ROC={auc_roc:.4f}')print('Best Hyperparameters:', best_hyperparameters)
print('Best Metric:', best_metric)inference.py 推理代码
import torch
import pandas as pd
from dataset_featurizer import MoleculeDataset
from sklearn.metrics import confusion_matrix, accuracy_score, roc_auc_score# Load the test dataset
test_dataset = MoleculeDataset(root="data/split_data", filename="HIV_test.csv", test=True)
test_loader = DataLoader(test_dataset, batch_size=NUM_GRAPHS_PER_BATCH, shuffle=True)# Load the trained model
model = torch.load(os.join.path(output_folder,"model.pth"))
model.eval()# Create lists to store the predicted and true labels
all_preds = []
all_labels = []
all_preds_raw = []# Perform inference on the test dataset
with torch.no_grad():for batch in test_loader:# Move the batch to the devicebatch = batch.to(device)# Perform forward passpred = model(batch.x.float(), batch.edge_attr.float(), batch.edge_index, batch.batch)# Convert the predictions to class labelspreds = torch.argmax(pred, dim=1)# Append the predicted and true labels to the listsall_preds.extend(preds.cpu().detach().numpy())all_labels.extend(batch.y.cpu().detach().numpy())all_preds_raw.extend(pred.cpu().detach().numpy())# Calculate the confusion matrix
cm = confusion_matrix(all_labels, all_preds)# Calculate the accuracy score
accuracy = accuracy_score(all_labels, all_preds)# Calculate the ROC AUC score
roc_auc = roc_auc_score(all_labels, all_preds_raw)# Print the confusion matrix, accuracy score, and ROC AUC score
print("Confusion Matrix:")
print(cm)
print("Accuracy Score:", accuracy)
print("ROC AUC Score:", roc_auc)config.py
import numpy as npHYPERPARAMETERS = {"batch_size": [32, 128, 64],"learning_rate": [0.1, 0.05, 0.01, 0.001],"weight_decay": [0.0001, 0.00001, 0.001],"sgd_momentum": [0.9, 0.8, 0.5],"scheduler_gamma": [0.995, 0.9, 0.8, 0.5, 1],"pos_weight" : [1.0], "model_embedding_size": [8, 16, 32, 64, 128],"model_attention_heads": [1, 2, 3, 4],"model_layers": [3],"model_dropout_rate": [0.2, 0.5, 0.9],"model_top_k_ratio": [0.2, 0.5, 0.8, 0.9],"model_top_k_every_n": [0],"model_dense_neurons": [16, 128, 64, 256, 32]
}'''
BEST_PARAMETERS = {"batch_size": [128],"learning_rate": [0.01],"weight_decay": [0.0001],"sgd_momentum": [0.8],"scheduler_gamma": [0.8],"pos_weight": [1.3],"model_embedding_size": [64],"model_attention_heads": [3],"model_layers": [4],"model_dropout_rate": [0.2],"model_top_k_ratio": [0.5],"model_top_k_every_n": [1],"model_dense_neurons": [256]
}'''