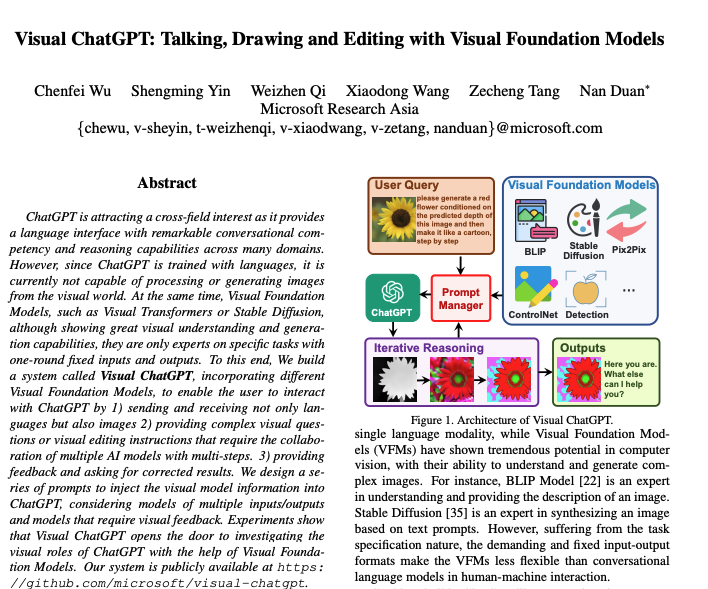
##绘制载荷图##
gene_loading <- as.data.frame(fit0$W.)
meta_loading <- as.data.frame(fit0$C.)
colnames(gene_loading) <- c("pq1","pq2")
colnames(meta_loading) <- c("pq1","pq2")
#添加新的一列,按组学进行归类#
Omics <- NA
gene_loading <- cbind(gene_loading[,1:2],Omics)
gene_loading[,3] <- "Transcriptome"
meta_loading <- cbind(meta_loading[,1:2],Omics)
meta_loading[,3] <- "Metabolome"
#两组数据合并#
mgloading <- rbind(meta_loading,gene_loading)
#绘图#
pp <- ggplot(data = mgloading,mapping = aes(x=pq1,y=pq2,colour=Omics))
pp

pp+geom_point()

pp <- ggplot(data = mgloading,mapping = aes(x=pq1,y=pq2,colour=Omics,shape=Omics,fill=Omics))
pp+geom_point()
pp+geom_point()+scale_color_manual(values=c("green","blue")) #更改颜色#

pp+geom_point(size=2)+scale_color_manual(values=c("green","blue"))+xlab("pq[1]")+ylab("pq[2]")+labs(title="O2PLS_loadings")+theme_bw()#切换成黑白背景的主题,添加主标题#

library(eoffice)
topptx(filename = "O2PLS_loadings.pptx")#导出到ppt#
參考:【功能代谢组学】O2PLS模型关联分
重磅O2PLS for Multi-Omics