目录
- 报错
- 解决办法
报错
笔者在使用 nnunetv2 进行 KiTS19肾脏肿瘤分割实验的训练步骤中
使用 2d 和3d_lowres 训练都没有问题
nnUNetv2_train 40 2d 0
nnUNetv2_train 40 3d_lowres 0
但是使用 3d_cascade_fullres 和 3d_fullres 训练
nnUNetv2_train 40 3d_cascade_fullres 0
nnUNetv2_train 40 3d_fullres 0
都会报这个异常 ValueError: mmap length is greater than file size
具体报错内容如下:
root@autodl-container-fdb34f8e52-02177b7e:~# nnUNetv2_train 40 3d_cascade_fullres 0
Using device: cuda:0#######################################################################
Please cite the following paper when using nnU-Net:
Isensee, F., Jaeger, P. F., Kohl, S. A., Petersen, J., & Maier-Hein, K. H. (2021). nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nature methods, 18(2), 203-211.
#######################################################################This is the configuration used by this training:
Configuration name: 3d_cascade_fullres{'data_identifier': 'nnUNetPlans_3d_fullres', 'preprocessor_name': 'DefaultPreprocessor', 'batch_size': 2, 'patch_size': [128, 128, 128], 'median_image_size_in_voxels': [525.5, 512.0, 512.0], 'spacing': [0.78126, 0.78125, 0.78125], 'normalization_schemes': ['CTNormalization'], 'use_mask_for_norm': [False], 'UNet_class_name': 'PlainConvUNet', 'UNet_base_num_features': 32, 'n_conv_per_stage_encoder': [2, 2, 2, 2, 2, 2], 'n_conv_per_stage_decoder': [2, 2, 2, 2, 2], 'num_pool_per_axis': [5, 5, 5], 'pool_op_kernel_sizes': [[1, 1, 1], [2, 2, 2], [2, 2, 2], [2, 2, 2], [2, 2, 2], [2, 2, 2]], 'conv_kernel_sizes': [[3, 3, 3], [3, 3, 3], [3, 3, 3], [3, 3, 3], [3, 3, 3], [3, 3, 3]], 'unet_max_num_features': 320, 'resampling_fn_data': 'resample_data_or_seg_to_shape', 'resampling_fn_seg': 'resample_data_or_seg_to_shape', 'resampling_fn_data_kwargs': {'is_seg': False, 'order': 3, 'order_z': 0, 'force_separate_z': None}, 'resampling_fn_seg_kwargs': {'is_seg': True, 'order': 1, 'order_z': 0, 'force_separate_z': None}, 'resampling_fn_probabilities': 'resample_data_or_seg_to_shape', 'resampling_fn_probabilities_kwargs': {'is_seg': False, 'order': 1, 'order_z': 0, 'force_separate_z': None}, 'batch_dice': True, 'inherits_from': '3d_fullres', 'previous_stage': '3d_lowres'}These are the global plan.json settings:{'dataset_name': 'Dataset040_KiTS', 'plans_name': 'nnUNetPlans', 'original_median_spacing_after_transp': [3.0, 0.78125, 0.78125], 'original_median_shape_after_transp': [108, 512, 512], 'image_reader_writer': 'SimpleITKIO', 'transpose_forward': [2, 0, 1], 'transpose_backward': [1, 2, 0], 'experiment_planner_used': 'ExperimentPlanner', 'label_manager': 'LabelManager', 'foreground_intensity_properties_per_channel': {'0': {'max': 3071.0, 'mean': 102.5714111328125, 'median': 103.0, 'min': -1015.0, 'percentile_00_5': -75.0, 'percentile_99_5': 295.0, 'std': 73.64986419677734}}}2023-10-13 17:22:36.747343: unpacking dataset...
2023-10-13 17:22:40.991390: unpacking done...
2023-10-13 17:22:40.992978: do_dummy_2d_data_aug: False
2023-10-13 17:22:40.997410: Using splits from existing split file: /root/autodl-tmp/nnUNet-master/dataset/nnUNet_preprocessed/Dataset040_KiTS/splits_final.json
2023-10-13 17:22:40.998125: The split file contains 5 splits.
2023-10-13 17:22:40.998262: Desired fold for training: 0
2023-10-13 17:22:40.998355: This split has 168 training and 42 validation cases.
/root/miniconda3/lib/python3.10/site-packages/torch/onnx/symbolic_helper.py:1513: UserWarning: ONNX export mode is set to TrainingMode.EVAL, but operator 'instance_norm' is set to train=True. Exporting with train=True.warnings.warn(
2023-10-13 17:22:45.383066:
2023-10-13 17:22:45.383146: Epoch 0
2023-10-13 17:22:45.383244: Current learning rate: 0.01
Exception in background worker 4:mmap length is greater than file size
Traceback (most recent call last):File "/root/miniconda3/lib/python3.10/site-packages/batchgenerators/dataloading/nondet_multi_threaded_augmenter.py", line 53, in produceritem = next(data_loader)File "/root/miniconda3/lib/python3.10/site-packages/batchgenerators/dataloading/data_loader.py", line 126, in __next__return self.generate_train_batch()File "/root/autodl-tmp/nnUNet-master/nnunetv2/training/dataloading/data_loader_3d.py", line 19, in generate_train_batchdata, seg, properties = self._data.load_case(i)File "/root/autodl-tmp/nnUNet-master/nnunetv2/training/dataloading/nnunet_dataset.py", line 86, in load_casedata = np.load(entry['data_file'][:-4] + ".npy", 'r')File "/root/miniconda3/lib/python3.10/site-packages/numpy/lib/npyio.py", line 429, in loadreturn format.open_memmap(file, mode=mmap_mode,File "/root/miniconda3/lib/python3.10/site-packages/numpy/lib/format.py", line 937, in open_memmapmarray = numpy.memmap(filename, dtype=dtype, shape=shape, order=order,File "/root/miniconda3/lib/python3.10/site-packages/numpy/core/memmap.py", line 267, in __new__mm = mmap.mmap(fid.fileno(), bytes, access=acc, offset=start)
ValueError: mmap length is greater than file size
Exception in background worker 2:mmap length is greater than file size
Traceback (most recent call last):File "/root/miniconda3/lib/python3.10/site-packages/batchgenerators/dataloading/nondet_multi_threaded_augmenter.py", line 53, in produceritem = next(data_loader)File "/root/miniconda3/lib/python3.10/site-packages/batchgenerators/dataloading/data_loader.py", line 126, in __next__return self.generate_train_batch()File "/root/autodl-tmp/nnUNet-master/nnunetv2/training/dataloading/data_loader_3d.py", line 19, in generate_train_batchdata, seg, properties = self._data.load_case(i)File "/root/autodl-tmp/nnUNet-master/nnunetv2/training/dataloading/nnunet_dataset.py", line 86, in load_casedata = np.load(entry['data_file'][:-4] + ".npy", 'r')File "/root/miniconda3/lib/python3.10/site-packages/numpy/lib/npyio.py", line 429, in loadreturn format.open_memmap(file, mode=mmap_mode,File "/root/miniconda3/lib/python3.10/site-packages/numpy/lib/format.py", line 937, in open_memmapmarray = numpy.memmap(filename, dtype=dtype, shape=shape, order=order,File "/root/miniconda3/lib/python3.10/site-packages/numpy/core/memmap.py", line 267, in __new__mm = mmap.mmap(fid.fileno(), bytes, access=acc, offset=start)
ValueError: mmap length is greater than file size
using pin_memory on device 0
Traceback (most recent call last):File "/root/miniconda3/bin/nnUNetv2_train", line 8, in <module>sys.exit(run_training_entry())File "/root/autodl-tmp/nnUNet-master/nnunetv2/run/run_training.py", line 268, in run_training_entryrun_training(args.dataset_name_or_id, args.configuration, args.fold, args.tr, args.p, args.pretrained_weights,File "/root/autodl-tmp/nnUNet-master/nnunetv2/run/run_training.py", line 204, in run_trainingnnunet_trainer.run_training()File "/root/autodl-tmp/nnUNet-master/nnunetv2/training/nnUNetTrainer/nnUNetTrainer.py", line 1237, in run_trainingtrain_outputs.append(self.train_step(next(self.dataloader_train)))File "/root/miniconda3/lib/python3.10/site-packages/batchgenerators/dataloading/nondet_multi_threaded_augmenter.py", line 196, in __next__item = self.__get_next_item()File "/root/miniconda3/lib/python3.10/site-packages/batchgenerators/dataloading/nondet_multi_threaded_augmenter.py", line 181, in __get_next_itemraise RuntimeError("One or more background workers are no longer alive. Exiting. Please check the "
RuntimeError: One or more background workers are no longer alive. Exiting. Please check the print statements above for the actual error message
解决办法
nnunet 作者给出的解决办法,详情请戳
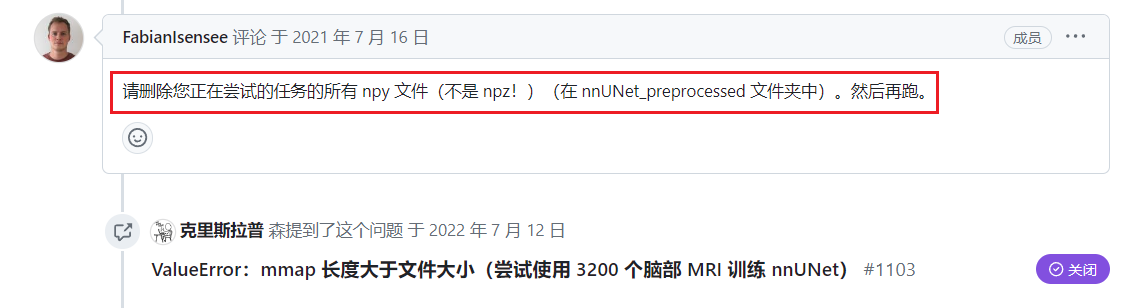
进入指定文件夹中,执行
rm *.npy