信息来源:
https://www.fda.gov/industry/fda-data-standards-advisory-board/study-data-standards-resources
STUDY DATA TECHNICAL CONFORMANCE GUIDE v4.9 (March 2022) (研究数据技术一致性指南)
仅提取该文档中的部分内容加以翻译,以下中文都是机器翻译,仅供自学
2.2 Study Data Reviewer’s Guides 临床数据评审指南
The preparation of the relevant Reviewer Guides (RG) is recommended as an integral part of a standards-compliant study data submission. An RG should describe any special considerations or directions or conformance issues that may facilitate an FDA reviewer's use of the submitted data and may help the reviewer understand the relationships between the study report and the data.
建议将相关审评员指南(RG)的编制作为符合标准的研究数据提交的一个组成部分。RG应描述任何特殊考虑因素或指示或一致性问题,这些问题可能有助于FDA审查员使用提交的数据,并可能有助于审查员了解研究报告与数据之间的关系。
There are two study data reviewer guides (SDRG): clinical and nonclinical. The SDRG for nonclinical studies (nSDRG) and SDRG for clinical studies (cSDRG) should be placed with the study data in Module 4 and 5, respectively, in the eCTD The SDRG should be file-tagged as ‘data-tabulation-data-definition’, with a clear leaf title.
有两种研究数据审评员指南(SDRG):临床和非临床。用于非临床研究的SDRG(nSDRG)和用于临床研究的SDRG(cSDRG)应分别与eCTD中的模块4和5中的研究数据一起放置SDRG的文件标记为“数据制表-数据定义”,并具有明确的叶子标题。
2.2.1 SDRG for Clinical Data 临床研究的SDRG
An SDRG for clinical data should be named cSDRG (the prefix ‘c’ designates ‘clinical’) and the document should be named ‘csdrg’ and provided as a PDF file upon submission (csdrg.pdf).
临床数据的SDRG应命名为cSDRG(前缀“c”表示“临床”),文档应命名为“csdrg”,并在提交时以PDF文件的形式提供(csdrg.pdf)。
2.3 Analysis Data Reviewer’s Guide 分析数据评审指南
The preparation of an Analysis Data Reviewer’s Guide (ADRG) is recommended as an important part of a standards-compliant analysis data submission for clinical trials. The ADRG provides FDA reviewers with context for analysis datasets and terminology, received as part of a regulatory product submission, additional to what is presented within the data folder (i.e., define.xml). The ADRG also provides a summary of ADaM conformance findings. The ADRG purposefully duplicates limited information found in other submission documentation (e.g., the protocol, statistical analysis plan (SAP), clinical study report, define.xml) in order to provide FDA reviewers with a single point of orientation to the analysis datasets. It should be noted that the submission of an ADRG does not eliminate the requirement to submit a complete and informative define.xml file corresponding to the analysis datasets.
建议将编制分析数据审查员指南 (ADRG) 作为临床试验符合标准的分析数据提交的重要组成部分。ADRG为FDA审查员提供了分析数据集和术语的上下文,这些背景是作为监管产品提交的一部分收到的,除了数据文件夹中显示的内容(define.xml)。ADRG 还提供了 ADaM 一致性结果的摘要。ADRG有目的地复制其他提交文档(例如,方案,统计分析计划(SAP),临床研究报告,define.xml)中发现的有限信息,以便为FDA审查员提供分析数据集的单一定位点。应该注意的是,提交ADRG并不能消除提交与分析数据集相对应的完整且信息丰富的define.xml文件的要求。
• The ADRG for a clinical study should be placed with the analysis data in Module 5 of the eCTD. The ADRG should be file-tagged as ‘analysis-data-definition’, with a clear leaf title.
用于临床研究的ADRG应与eCTD模块5中的分析数据一起放置。ADRG应将文件标记为“分析数据定义”,并带有明确的叶子标题。
• An ADRG for clinical data should be called an ADRG and the document should be a PDF file ‘adrg.pdf’ upon submission.
用于临床数据的ADRG应称为ADRG,提交时文档应为PDF文件“adrg.pdf”。
3. Exchange Format – Electronic Submissions 交换格式 – 电子提交
3.1 Extensible Mark-up Language 可扩展标记语言
Extensible Mark-up Language (XML), as defined by the World Wide Web Consortium (W3C), specifies a set of rules for encoding documents in a format that is both humanreadable and machine-readable XML facilitates the sharing of structured data across different information systems. An XML use case is CDISC’s define.xml file. All XML files should use .xml as the file extension. Although XML files can be compressed, the define.xml should not be compressed.
由万维网联合会 (W3C) 定义的可扩展标记语言 (XML) 指定了一组规则,用于以人类可读和机器可读的 XML 格式对文档进行编码,从而有助于在不同信息系统之间共享结构化数据。XML 用例是 CDISC 的define.xml文件。所有 XML 文件都应使用.xml作为文件扩展名。尽管 XML 文件可以压缩,但不应压缩define.xml。
3.2 Portable Document Format 可移植文档格式
Portable Document Format (PDF) is an open file format used to represent documents in a
manner independent of application software, hardware, and operating systems. A PDF use case includes, e.g., the annotated CRF (aCRF / blank crf), and other documents that align with the International Council for Harmonisation (ICH) M2 FDA PDF specifications are located on FDA’s eCTD Web site. The Catalog lists the PDF version(s) that are supported by FDA. All PDF files should use .pdf as the file extension.
可移植文档格式 (PDF) 是一种开放文件格式,用于以独立于应用程序软件、硬件和操作系统的方式表示文档。PDF用例包括,例如,带注释的CRF(aCRF / 空白crf),以及符合国际协调委员会(ICH)M2 FDA PDF规范的其他文档位于FDA的eCTD网站上。该目录列出了FDA支持的PDF版本。所有PDF文件都应使用.pdf作为文件扩展名。
3.3 File Transport Format 文件传输格式
3.3.1 SAS Transport Format SAS传输格式
The SAS Transport Format (XPORT) Version 5 is the file format for the submission of all electronic datasets.The XPORT is an open file format published by SAS Institute for the exchange of study data. Data can be translated to and from XPORT to other commonly used formats without the use of programs from SAS Institute or any specific vendor. There should be one dataset per transport file, and the dataset in the transport file should be named the same as the transport file (e.g., ‘ae’ and ae.xpt, ‘suppae’ and suppae.xpt, ‘lb1’ and lb1.xpt).
SAS 传输格式 (XPORT) 版本 5 是用于提交所有电子数据集的文件格式。XPORT 是由 SAS 研究所发布的用于交换研究数据的开放文件格式。数据可以在XPORT之间来回转换到其他常用格式,而无需使用SAS Institute或任何特定供应商的程序。每个传输文件应该有一个数据集,并且传输文件中的数据集应与传输文件的名称相同(例如,'ae'和ae.xpt,'suppae'和suppae.xpt,'lb1'和lb1.xpt)。
XPORT files can be created by the COPY Procedure in SAS Version 5 and higher of the
SAS Software. SAS Transport files processed by the SAS CPORT cannot be reviewed,
processed, or archived by FDA. Sponsors can find the record layout for SAS XPORT
transport files through SAS technical document TS-140. All SAS XPORT transport
files should use .xpt as the file extension. There should be one dataset per XPORT file,
and the files should not be compressed.
XPORT文件可以通过SAS软件的SAS版本5及更高版本中的复制过程创建。FDA 无法审核、处理或存档由 SAS CPORT 处理的 SAS 传输文件。赞助商可以通过SAS技术文档TS-140找到SAS XPORT传输文件的记录布局。所有 SAS XPORT 传输文件都应使用 .xpt 作为文件扩展名。每个 XPORT 文件应该有一个数据集,并且不应压缩这些文件。
3.3.2 Dataset Size 数据集大小
Each dataset should be provided in a single transport file. The maximum size of an individual dataset that FDA can process depends on many factors. Datasets greater than 5 gigabytes (GB) in size should be split into smaller datasets no larger than 5 GB. Sponsors should submit these smaller datasets, in addition to the larger non-split datasets, to better support regulatory reviewers. The split datasets should be placed in a separate subdirectory labeled ‘split’ (See section 7.2). A clear explanation regarding how these datasets were split needs to be presented within the relevant data RG.
每个数据集都应在单个传输文件中提供。FDA可以处理的单个数据集的最大大小取决于许多因素。大小大于 5千兆字节 (GB) 的数据集应拆分为不大于 5 GB 的较小数据集。申办方除了较大的非拆分数据集外,还应提交这些较小的数据集,以更好地支持监管审查员。拆分数据集应放置在标记为“split”的单独子目录中(请参阅第 7.2 节)。关于这些数据集是如何拆分的清晰解释需要在相关数据RG中呈现。
3.3.3 Dataset Column Length 数据集列长度
The allotted length for each column containing character (text) data should be set to the maximum length of the variable used across all datasets in the study except for suppqual datasets. For suppqual datasets, the allotted length for each column containing character (text) data should be set to the maximum length of the variable used in the individual dataset. This will significantly reduce file sizes. For example, if USUBJID has a maximum length of 18, the USUBJID’s column size should be set to 18, not 200. Care should be taken to avoid accidental truncation of data through dataset merges. Ensure that variable length reduction happens before datasets are split. For example, if PARAM is set to a length of 20 in ADLB1 and 25 in ADLB2, when ADLB2 is concatenated with ADLB1 data loss will occur. SAS uses the length of 20 for the width which will truncate data in ADLB2 when the contents of the PARAM field is longer than 20 characters.
包含字符(文本)数据的每列的分配长度应设置为研究中除 suppqual数据集之外的所有数据集中使用的变量的最大长度。对于替代数据集,为包含字符(文本)数据的每列分配的长度应设置为单个数据集中使用的变量的最大长度。这将显著减小文件大小。例如,如果 USUBJID 的最大长度为 18,则 USUBJID 的列大小应设置为 18,而不是 200。[译注:Pinnacle 21可以报告该最大长度问题] 应注意以避免通过数据集合并意外截断数据。确保在拆分数据集之前进行可变长度减少。例如,如果 PARAM 在 ADLB1 中设置为 20,在 ADLB2 中设置为 25,则当 ADLB2与 ADLB1 串联时,将发生数据丢失。SAS 使用长度 20 作为宽度,当 PARAM 字段的内容长度超过 20 个字符时,这将截断 ADLB2 中的数据。
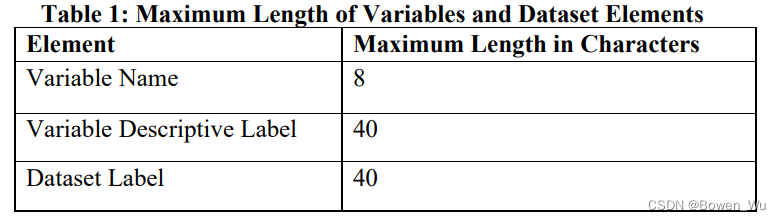
3.3.4 Variable and Dataset Descriptor Length 变量和数据集描述符长度
The length of variable names, descriptive labels, and dataset labels should not exceed the maximum permissible number of characters described in Table 1
变量名称、描述性标签和数据集标签的长度不应超过表 1 中描述的最大允许字符数
3.3.5 Special Characters: Variables and Datasets 特殊字符:变量和数据集
Variable names, as well as variable and dataset labels should include American Standard Code for Information Interchange (ASCII) text codes only. Variable values are the most broadly compatible with software and operating systems when they are restricted to ASCII text codes (printable values below 128). Use UTF-8 for extending character sets; however, the use of extended mappings is not recommended. Transcoding errors, variable length errors, and lack of software support for multi byte UTF-8 encodings can result in incorrect character display and variable value truncations. Ensure that LBSTRESC and controlled terminology extensions in LBTEST do not contain byte values 160-191 as some character mappings in that range may interfere with agency processes.
变量名称以及变量和数据集标签应仅包含美国信息交换标准代码 (ASCII) 文本代码。当变量值仅限于 ASCII 文本代码(可打印值低于 128)时,它们与软件和操作系统最广泛兼容。使用 UTF-8 扩展字符集;但是,不建议使用扩展映射。转码错误、可变长度错误以及缺少对多字节 UTF-8 编码的软件支持可能会导致不正确的字符显示和可变值截断。确保 LBSTRESC 和 LBTEST 中的受控术语扩展不包含字节值 160-191 [译注:扩展字节的编码见ASCII Code - The extended ASCII table],因为该范围内的某些字符映射可能会干扰代理流程。
3.3.6 Variable and Dataset Names 变量和数据集名称
Variable names should contain only uppercase letters, numbers, and must start with a letter. Dataset names should contain only lowercase letters, numbers, and must start with a letter. No other symbols or special characters should be included in these names. For legacy studies started on or before December 17, 2016, it is permissible to use the underscore character _ in variable names and dataset names.
变量名称应仅包含大写字母、数字,并且必须以字母开头。数据集名称应仅包含小写字母、数字,并且必须以字母开头。这些名称中不应包含其他符号或特殊字符。对于在 2016 年 12 月 17 日或之前开始的旧版研究,允许在变量名称和数据集名称中使用下划线字符 _ 。
3.3.7 Variable and Dataset Labels 变量和数据集标签
Do not include the following special characters in variable and dataset labels:
1/ Unbalanced apostrophe, e.g., “Parkinson's”
2/ Unbalanced single and double quotation marks
3/ Unbalanced parentheses, braces or brackets, e.g.,‘(‘, ‘{‘and ‘[‘
不要在变量和数据集标签中包含以下特殊字符:
1/ 不配对的撇号,例如“Parkinson's”
2/ 不配对的单引号和双引号
3/ 不配对的括号、大括号或方括号,例如,“(”、“{” 和 “[”
4. Study Data Submission Format – Clinical and Nonclinical 研究数据提交格式 – 临床和非临床
4.1 Clinical Data Interchange Standards Consortium 临床数据交换标准协会
Clinical Data Interchange Standards Consortium (CDISC) is an open, multidisciplinary, neutral, nonprofit standards development organization (SDO) that has been working through consensus-based collaborative teams to develop global data standards for clinical and nonclinical research
临床数据交换标准联盟 (CDISC) 是一个开放、多学科、中立、非营利性标准制定组织 (SDO),它一直通过基于共识的协作团队为临床和非临床研究开发全球数据标准。
Data format specifications for the tabulation datasets of clinical and nonclinical toxicology studies are provided by SDTM and SEND, respectively, while data format specifications for the analysis datasets of clinical studies are provided by ADaM. It should be noted that data format specifications for the analysis datasets of nonclinical toxicology studies have not been developed. As noted in section 1.1, the Catalog provides a listing of the currently supported data standards with links to reference materials. For the purposes of this Guide, the terms SDTM, ADaM, and SEND apply to versions only listed and supported by FDA in the Catalog.
临床和非临床毒理学研究的制表数据集的数据格式规范分别由SDTM和SEND提供,而临床研究分析数据集的数据格式规范由ADaM提供。应该指出的是,非临床毒理学研究的分析数据集的数据格式规范尚未制定。如第1.1节所述,该目录提供了当前支持的数据标准的清单,并附有参考资料的链接。就本指南而言,术语SDTM、ADaM和SEND仅适用于FDA在目录中列出和支持的版本。
Although the SDTM and SEND formats facilitate review of the data, they do not always provide the data structured in a way that supports all analyses needed for review. Analysis files are critical for FDA to understand, on a per subject basis, how the specific analyses contained in the study report have been created. Therefore, sponsors should supplement the SDTM with ADaM analysis datasets as described below.
尽管SDTM和SEND格式有助于对数据进行审查,但它们并不总是以支持审查所需的所有分析的方式提供结构化的数据。因此,申办方应使用 ADaM 分析数据集来补充 SDTM,如下所述。
There may be instances in which current implementation guides (e.g., SDTMIG, SENDIG) do not provide specific instruction as to how certain study data should be represented. In these instances, sponsors should discuss their proposed solution with the review division and submit supporting documentation that describes these decisions or solutions in the appropriate SDRG at the time of submission.
在有些情况下,目前的实施指南(例如,SDTMIG,SENDIG)没有就如何表示某些研究数据提供具体说明。在这些情况下,申办者应与评审部门讨论他们提出的解决方案,并在提交时在适当的SDRG中提交描述这些决定或解决方案的支持文档。
4.1.1 Study Data Tabulation Model 研究数据列表模型
4.1.1.1 Definition 定义
The Study Data Tabulation Model (SDTM) defines a standard structure for human clinical trials tabulation datasets
研究数据制表模型(SDTM)定义了人体临床试验制表数据集的标准结构
4.1.1.2 SDTM General Considerations SDTM一般注意事项
It is recommended that sponsors implement the SDTM standard for representation of clinical trial tabulation data prior to the conduct of the study.
建议申办者在进行研究前实施SDTM标准,以表示临床试验制表数据。
The SDTMIG should be followed unless otherwise indicated in this Guide or in the Catalog. The conformance criteria listed in the SDTMIG should not be interpreted as the sole determinant of the adequacy of submitted data. If there is uncertainty regarding implementation, the sponsor should discuss application-specific questions with the review division and general standards implementation questions with the specific center resources identified elsewhere in this Guide (See section 1.2). Each submitted SDTM dataset should have its contents described with complete metadata in the define.xml file (See section 4.1.4.5) and within the cSDRG as appropriate (See section 2.2). When updated datasets (e.g., ‘ae.xpt’, ‘lb.xpt’) are submitted, updated and complete define.xml and cSDRG covering all datasets should be submitted using the “replace” lifecycle operator to update the original file.
除非本指南([译注:指该FDA指南])或目录中另有说明,否则应遵循SDTMIG。SDTMIG中列出的一致性标准不应被解释为所提交数据充分性的唯一决定因素。如果实施存在不确定性,申办方应与审查部门讨论特定于应用的问题,并与本指南其他地方确定的特定中心资源讨论一般标准实施问题(见第1.2节)。每个提交的 SDTM 数据集都应在define.xml文件(请参阅第 4.1.4.5 节)和 cSDRG(请参阅第 2.2 节)中用完整的元数据描述其内容。当提交更新的数据集(例如,'ae.xpt','lb.xpt')时,应使用“替换”生命周期运算符提交更新和完整的define.xml和涵盖所有数据集的cSDRG应使用“替换”生命周期运算符来更新原始文件。
Except for variables that are defined in the SDTMIG as being coded, numerically coded variables typically are not submitted as part of the SDTM datasets. Numeric values generated from validated scoring instruments or questionnaires do not represent codes, and therefore have no relevance for this issue. There may be special instances when codes are preferred, hence sponsors should refer to the review division for direction, if there are any questions.
除了在 SDTMIG 中定义为已编码的变量外,数字编码的变量通常不作为 SDTM 数据集的一部分提交。从经过验证的评分工具或调查表生成的数值不表示代码,因此与此问题无关。在特殊情况下,代码是首选的,因此,如果有任何疑问,申办方应向审查部门寻求指导。
Subject Identifier (SUBJID)
The variable SUBJID uniquely identifies each subject that participates in a study. If a single subject is screened and/or enrolled more than once in a study, then the subject’s SUBJID should be different for each unique screening or enrollment. For a study with multiple screenings and/or multiple enrollments per subject, SUBJID should be included in other related domains besides DM even though it may cause validation errors. It is recommended to include a table linking each SUBJID for a single subject to that subject’s USUBJID with any additional necessary explanation included in the relevant RG.
变量 SUBJID 唯一标识参与研究的每个受试者。如果单个受试者在研究中被筛选和/或注册多次,则对于每个独特的筛选或入组,受试者的SUBJID应该不同。对于每个受试者进行多次筛选和/或多次入组的研究,SUBJID应包含在DM以外的其他相关领域中,即使它可能导致验证错误。[译注:SDTM IG 目前v3.3版本中没有关于该情况的处理规则,申办方为了满足FDA此要求,通常会创建新的custom domain, 比如XM或WD 来储存rescreening的record]。 建议包括一个表格,将单个主题的每个SUBJID链接到该主题的USUBJID,并在相关RG中包含的任何其他必要解释。
Unique Subject Identifier (USUBJID)
The variable USUBJID is an identifier used to uniquely identify a subject across all studies for all applications or submissions involving the product. Each individual subject should be assigned a single unique identifier across the entire application. This is in addition to the subject ID (SUBJID) used to identify subjects in each study and its corresponding study report. An individual subject should have the exact same unique identifier across all datasets, including between SDTM and ADaM datasets. Subjects that participate in more than one study should maintain the same USUBJID across all studies. It is important to follow this convention to enable pooling of a single subject’s data across studies (e.g., a randomized control trial and an extension study).
变量 USUBJID 是一个标识符,用于在涉及Product的所有申请或提交的所有研究中唯一标识受试者。应在整个应用程序中为每个单独的主题分配一个唯一标识符。这是对用于识别每项研究中的受试者及其相应研究报告的受试者ID(SUBJID)的补充。单个受试者应在所有数据集(包括 SDTM 和 ADaM 数据集之间)具有完全相同的唯一标识符。参与多项研究的受试者应在所有研究中保持相同的USUBJID。遵循这一惯例对于跨研究(例如,随机对照试验和扩展研究)汇集单个受试者的数据非常重要。
Sponsors should not add leading or trailing spaces to the USUBJID variable in any dataset. For example, applications have been previously submitted in which the USUBJID variable for each individual subject appeared to be the same across datasets; however, in certain datasets, the actual entry had leading zeros added, or zeros added elsewhere in the entry. This does not allow for machine-readable matching of individual subject data across all datasets. Improper implementation of the USUBJID variable is a common error with applications and often requires sponsors to re-submit their data
申办方不应在任何数据集中向 USUBJID 变量添加前导空格或尾随空格。例如,以前提交的申请中,每个受试者的USUBJID变量在数据集中似乎都是相同的;但是,在某些数据集中,实际条目添加了前导零,或者在条目中的其他位置添加了零。这不允许在所有数据集中对单个主题数据进行机器可读匹配。USUBJID变量的不正确实现是应用程序的常见错误,通常需要赞助商重新提交其数据
Adjudication Data 裁决数据
There are no existing standards or best practices for the representation of adjudication data as part of a standard data submission. Until standards for adjudication data are developed, it is advised that sponsors discuss their proposed approach with the review division and also include details about the presence, implementation approach, and location of adjudication data in the SDRG.
没有关于将裁决数据表示为标准数据提交一部分的现有标准或最佳做法。在制定裁决数据标准之前,建议申办人与审查部门讨论他们提出的方法,并包括有关裁决数据在SDRG中的存在,实施方法和位置的详细信息。
4.1.1.3 SDTM Domain Specifications SDTM 域规范
SUPPQUAL (Supplemental Qualifier)
A SUPPQUAL dataset is a special SDTM dataset that contains non-standard variables which cannot be represented in the existing SDTM domains. SUPPQUAL should be used only when key data cannot be represented in SDTM domains. In general, variables used to support key analyses should not be represented in SUPPQUAL. Discussion with the review division should occur if the sponsor intends to include important variables (e.g., that support key analyses) in SUPPQUAL datasets, and this should be reflected in the SDRG.
SUPPQUAL 数据集是一种特殊的 SDTM 数据集,其中包含无法在现有 SDTM 域中表示的非标准变量。仅当关键数据无法在 SDTM 域中表示时,才应使用 SUPPQUAL。通常,用于支持关键分析的变量不应在 SUPPQUAL 中表示。如果申办者打算在SUPPQUAL数据集中包括重要变量(例如,支持关键分析),则应与审查部门进行讨论,这应反映在SDRG中。
DM Domain (Demographics)
In the DM domain, each subject should have only one single record per study. Screen failures, when provided, should be included as a record in DM with the ARM, ARMCD, ACTARM, and ACTARMCD field left blank. For subjects who are randomized in treatment group but not treated, the planned arm variables (ARM and ARMCD) should be populated, but actual treatment arm variables (ACTARM and ACTARMCD) should be left blank.
在DM领域,每个受试者每项研究应该只有一个记录。筛选失败(如果提供)应作为记录包含在 DM 中,并将 ARM、ARMCD、ACTARM 和 ACTARMCD 字段留空。[译注:SDTM IG v3.3中DM domain的更新已与此规则相适应;如果递交材料采用的是IG v3.2,则应注意此规则与IG v3.2中DM domain的假设有冲突之处] 对于随机分为治疗组但未接受治疗的受试者,应填充计划的手臂变量(ARM 和 ARMCD),但实际治疗组变量(ACTARM 和 ACTARMCD)应留空。
For subjects with multiple enrollments within a single study, the primary enrollment should be submitted in DM. Additional enrollments should be included in a custom domain with a similar structure to DM. Clarifying statements in the RG would be helpful.
对于在单个研究中有多次入组的受试者,主要入组应以DM格式提交。其他入组应包含在与DM结构相似的自定义域中。[译注:SDTM IG 目前v3.3版本中没有关于该情况的处理规则,申办方为了满足FDA此要求,通常会创建新的custom domain, 比如XM或WD 来储存重复入组的观测 澄清RG中的陈述会有所帮助。
For subjects with multiple screenings and no subsequent enrollment, include the primary screening in DM with additional screenings in a custom domain with a structure similar to DM.
对于多次筛查且没有后续入组的受试者,需要在DM中的包含初次筛查,以及在结构与DM相似的自定义域中的包含额外筛查。
For subjects with multiple screenings and subsequent enrollment, include the enrollment in DM with screenings in a custom domain with a structure similar to DM.
对于多次筛选和随后入组的受试者,需要在DM中包含入组信息,并在具有类似于DM的结构的自定义域中包含筛选信息。
DS Domain (Disposition)
When there is more than one disposition event, the EPOCH or DSSCAT variable should be used to aid in distinguishing between them. This will allow identification of the EPOCH in which each event occurred or DSSCAT to differentiate if the disposition is for treatment or study. If a death of any type occurs, it should be the last record and should include its associated EPOCH. It is expected that EPOCH variable values will be determined based on the trial design and thus should be defined clearly and documented in the define.xml.
当存在多个处置事件时,应使用 EPOCH 或 DSSCAT 变量来帮助区分它们。[译注:SDTM.IG 3.3 中DS域已新增假设与此规则适应] 这将允许识别每个事件发生的EPOCH或DSSCAT以区分处置是用于治疗还是研究。如果发生任何类型的死亡,它应该是最后一个记录,并应包括其相关的EPOCH。预计EPOCH变量值将根据试验设计确定,因此应明确定义并记录在define.xml中。
SE Domain (Subject Elements)
The Subject Elements domain should be included to aid in the association of subject data (e.g., findings, events, and interventions) with the study element in which they occurred.
应包含受试者元素域,以帮助将受试者数据(例如,发现、事件和干预措施)与发生它们的研究要素相关联。
AE Domain (Adverse Events)
The AE domain should include all adverse events, unless otherwise specified in Technical Specification Document(s) appropriate for the indication.The definition of treatment emergent adverse events should be agreed upon with the review division and specified in the protocol (e.g., any AE after first dose of investigational product administration, or any AE after first dose of investigational product administration until X days after the last dose).
AE域应包括所有不良事件,除非技术规范文档中另有规定,适用于该适应症。治疗紧急不良事件的定义应与评价部门商定,并在方案中具体说明(例如,在首次给药研究产品后的任何AE,或在第一剂研究产品给药后至最后一次给药后X天的任何AE)。
The entry of a ‘Y’ for the serious adverse event variable, AESER, should have the assessment indicated (e.g., as a death, hospitalization, or disability/permanent damage). Frequently, sponsors omit the assessment information, even when it has been collected on the CRF. The criteria that led to the determination should be provided. This information is critical during FDA review to support the characterization of serious AEs.
严重不良事件变量 AESER 输入“Y”时,应指明评估(例如,死亡、住院或残疾/永久性损伤)[译注:在PMDA的一些项目中,SDTM只收集AESER=Y这一信息,具体评估信息不会收集到CRF,或者收集到CRF但不会做到SDTM中,此为PMDA与FDA的差异?待考证。] 申办方经常忽略评估信息,即使这些信息是在通用报告格式上收集的。应提供导致确定的标准。在FDA审查期间,这些信息对于支持严重AE的表征至关重要。
Custom Domains
The SDTMIG permits the creation of custom domains if the data do not fit into an existing domain. Prior to creating a custom domain, sponsors should confirm that the data do not fit into an existing domain. If it is necessary to create custom domains, sponsors should follow the recommendations in the SDTMIG. In addition, sponsors should present their implementation approach in the cSDRG. To provide study data that do not fit into an existing SDTM domain or draft SDTM domain, consider creating a custom dataset aligned with the Study Data Tabulation Model (SDTM). Questions about custom domains should be addressed in pre-submission meetings and documented in the SDSP.
SDTMIG 允许在数据不适合现有域时创建自定义域。在创建自定义域之前,发起人应确认数据不适合现有域。如果需要创建自定义域,申办方应遵循SDTMIG中的建议。此外,申办者应在cSDRG中介绍其实施方法。要提供不适合现有 SDTM 域或草稿 SDTM 域的研究数据,请考虑创建与研究数据制表模型 (SDTM) 对齐的自定义数据集。有关自定义域的问题应在提交前会议中解决,并记录在SDSP中。
LB Domain (Laboratory)
The size of the LB domain dataset submitted by sponsors is often too large to process (See section 3.3.2). This issue can be addressed by splitting a large LB dataset into smaller datasets according to LBCAT and LBSCAT, using LBCAT for initial splitting. If the size is still too large, then use LBSCAT for further splitting. For example, use the dataset name lb1 (file name ‘lb1.xpt’) for chemistry, dataset name lb2 (file name ‘lb2.xpt’) for hematology, and dataset name lb3 (file name ‘lb3.xpt’) for urinalysis. Splitting the dataset in other ways (e.g., by subject or file size) makes the data less useable. Sponsors should submit these smaller files in addition to the larger non-split standard LB domain file. Sponsors should submit the split files in a separate subdirectory/split that is clearly documented in addition to the non-split standard LB domain file in the SDTM datasets directory (See section 7). FDA may require laboratory data using conventional units for reviewing submissions and labeling. Sponsors should discuss with the review divisions what laboratory data should utilize conventional units prior to submission.
申办方提交的 LB 域数据集的大小通常太大而无法处理(请参阅第 3.3.2 节)。此问题可以通过根据 LBCAT 和 LBSCAT 将大型 LB 数据集拆分为较小的数据集,并使用 LBCAT 进行初始拆分来解决。如果大小仍然太大,则使用LBSCAT进行进一步拆分。例如,使用数据集名称 lb1(文件名'lb1.xpt')进行化学分析,数据集名称lb2(文件名'lb2.xpt')用于血液学,数据集名称lb3(文件名'lb3.xpt')用于尿液分析。以其他方式(例如,按主题或文件大小)拆分数据集会使数据的可用性降低。除了较大的非拆分标准 LB 域文件外,申办方还应提交这些较小的文件。发起人应在单独的子目录/拆分中提交拆分文件,该子目录/拆分除了 SDTM 数据集目录中的非拆分标准 LB 域文件外,还会清楚地记录该子目录/拆分文件(请参阅第 7 节)。FDA可能需要使用常规单位的实验室数据来审查提交和标签。申办者应在提交之前与审查部门讨论应使用哪些实验室数据常规单位。
Trial Design Model (TDM)
Unless a simplified ts.xpt is indicated (see below), all TDM datasets should be included with each SDTM study submission to describe the planned conduct of a clinical study
除非指示简化的ts.xpt(见下文),否则所有TDM数据集都应包含在每个SDTM研究提交中,以描述临床研究的计划进行
When submitting a full ts.xpt, please refer to the appendix section for a list of study parameters that should be submitted where relevant for clinical studies. Additional parameters may be included beyond those listed in the appendix. For clinical studies, study start date (SSTDTC) is the earliest date of informed consent among any subject that enrolled in the study
提交完整的ts.xpt时,请参阅附录部分,了解在与临床研究相关的研究中应提交的研究参数列表。除附录中列出的参数外,还可以包括其他参数。对于临床研究,研究开始日期(SSTDTC)是参加研究的任何受试者中知情同意的最早日期
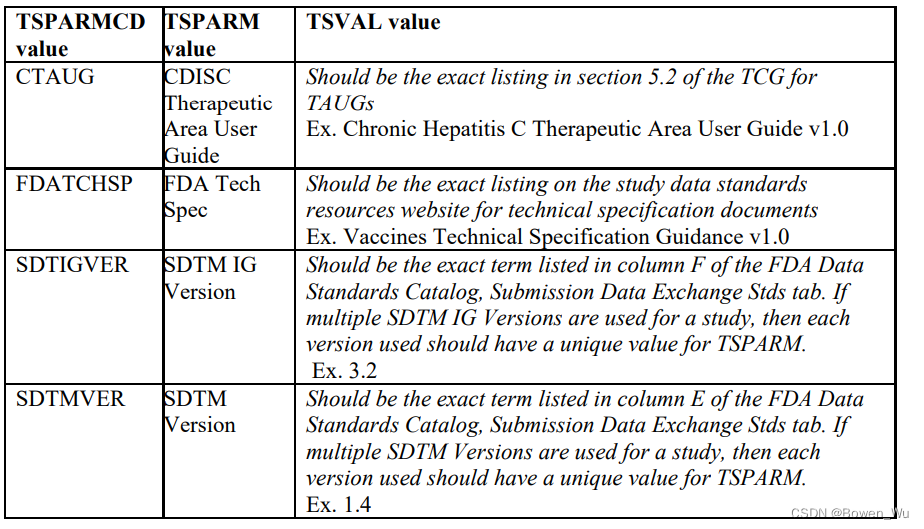
In addition to the study parameters indicated in the appendix section, if the study data submitted follows a Therapeutic Area User Guide (TAUG) or an FDA Technical Specification, use the values for TSPARM/TSPARMCD and TSVAL from the table below in the TS domain. Use of these parameters in TS will allow for tracking and reporting on the submission rates of study data following a particular TAUG or technical specification. At this time, it is also helpful to include the version of the CDISC implementation guide (IG) and model used using the parameters indicated in the table below.
除了附录部分中指示的研究参数外,如果提交的研究数据遵循治疗领域用户指南(TAUG)或FDA技术规范,请使用TS域中下表中的TSPARM / TSPARMCD和TSVAL的值。在TS中使用这些参数将允许跟踪和报告遵循特定TAUG或技术规范的研究数据的提交率。此时,包含 CDISC 实现指南 (IG) 的版本和使用下表中所示参数的模型也很有帮助。
EC Domain (Exposure as Collected)
The Exposure as Collected domain provides for protocol-specified study treatment administrations, as-collected. The EC domain may address some challenges in providing a subject’s exposure to study medication.
“作为收集的暴露”域提供方案指定的研究治疗管理,作为收集。EC领域可以解决在提供受试者接触研究药物方面的一些挑战。[译注:没明白这段要求的含义]
DD Domain (Death Details)
The Death Details domain provides for supplemental data that are typically collected when a death occurs, such as the official cause of death. The AE domain variables, AEOUT, AESDTH and AEENDTC/AEENDY should be populated and consistent with the death details.
“死亡详细信息”域提供通常在死亡发生时收集的补充数据,例如官方死因。应填充 AE 域变量 AEOUT、AESDTH 和 AEENDTC/AEENDY,并与死亡详细信息保持一致。
QS Domain (Questionnaires)
Some items in an instrument may be logically skipped per the instrument’s instructions. Responses for logically skipped items should be (1) recorded and/or scored according to the instructions provided in the instrument’s user manual, scoring manual, or other documentation provided by the instrument developer and (2) included in the submission dataset.
根据仪器的指令,仪器中的某些项目可能会在逻辑上跳过。逻辑跳过项目的响应应 (1) 根据仪器的用户手册、评分手册或仪器开发人员提供的其他文档中提供的说明进行记录和/或评分,以及 (2) 包含在提交数据集中。
If instructions on how to record and/or score responses to logically skipped items are available from the instrument developer, then records for logically skipped items should be included in the submission dataset with the following:
• QSSTAT = ‘NOT DONE’;
• QSREASND = ‘LOGICALLY SKIPPED ITEM’; and
• QSORRES, QSSTRESC, and QSSTRESN would be assigned according to the instrument’s instructions.
如果仪器开发人员提供了有关如何记录和/或评分对逻辑跳过项目的响应的说明,则逻辑跳过项目的记录应包含在提交数据集中,如下所示:
• QSSTAT = 'NOT DONE';
• QSREASND = '逻辑跳过项目';和
•QSORRES,QSSTRESC和QSSTRESN将根据仪器的指令进行分配。
If instructions on how to record and/or score responses to logically skipped items are not available from the instrument developer, then records for logically skipped items should be included in the submission dataset with the following:
• QSSTAT = ‘NOT DONE’;
• QSREASND = ‘LOGICALLY SKIPPED ITEM’; and
• QSORRES, QSSTRESC, and QSSTRESN all set to null.
如果仪器开发人员无法获得有关如何记录和/或评分对逻辑跳过项目的响应的说明,则逻辑跳过项目的记录应包含在提交数据集中,如下所示:
• QSSTAT = 'NOT DONE';
• QSREASND = '逻辑跳过的项目';和
• QSORRES、QSSTRESC 和 QSSTRESN 都设置为 null。
DV Domain (Protocol Deviations)
The DV domain should be included in your submission. It will be used by reviewers to examine protocol deviation trends of various study sites in order to facilitate the Bioresearch Monitoring Program (BIMO) clinical investigator site selection process, and once FDA tools are developed to extract and format needed data from SDTM, to populate line listings used by the Office of Regulatory Affairs (ORA) investigators during inspections. The following variables besides CDISC required variables should be included in the DV domain when submitting DV data: DVSPID, DVTERM, DVDECOD, DVCAT, DVSCAT, DVSTDTC, DVENDTC and EPOCH.
DV 域应包含在你的提交中。审稿人将使用它来检查各个研究站点的方案偏差趋势,以促进生物研究监测计划(BIMO)临床研究者选址过程,并且一旦开发了FDA工具来从SDTM中提取和格式化所需的数据,以填充监管事务办公室(ORA)研究人员在检查期间使用的线路列表。提交 DV 数据时,除了 CDISC 必需的变量外,以下变量还应包含在 DV 域中:DVSPID、DVTERM、DVDECOD、DVCAT、DVSCAT、DVSTDTC、DVENDTC 和 EPOCH。
4.1.2 Analysis Data Model 分析数据模型
4.1.2.1 Definition 定义
Specifications for analysis datasets for human drug product clinical studies are provided by the Analysis Data Model (ADaM) and its implementation by the Analysis Data Model Implementation Guide (ADaMIG). ADaM datasets should be used to create and to support the results in clinical study reports (CSRs), Integrated Summaries of Safety (ISS), and Integrated Summaries of Efficacy (ISE), as well as other analyses required for a thorough regulatory review. ADaM datasets can contain imputed data or data derived from SDTM datasets
人类药物产品临床研究分析数据集的规范由分析数据模型(ADaM)提供,其实施由分析数据模型实施指南(ADaMIG)提供。ADaM数据集应用于创建和支持临床研究报告(CSR),综合安全性摘要(ISS)和综合疗效摘要(ISE)中的结果,以及彻底监管审查所需的其他分析。ADaM 数据集可以包含插补数据或从 SDTM 数据集派生的数据
4.1.2.2 General Considerations 一般注意事项
Generally, ADaM assists FDA review. However, it does not always provide data structured in a way that supports all of the analyses that should be submitted for review. For example, ADaM structures do not support simultaneous analysis of multiple dependent variables or correlation analysis across several response variables. Therefore, sponsors should, as needed, supplement their ADaM datasets after discussions with the specific review division.
通常,ADaM协助FDA审查。但是,它并不总是以支持应提交审核的所有分析的方式提供结构化数据。例如,ADaM 结构不支持同时分析多个因变量或跨多个响应变量进行相关性分析。因此,申办者应在与具体审查部门讨论后,根据需要补充其ADaM数据集。
One of the expected benefits of analysis datasets that conform to ADaM is that they simplify the
programming steps necessary for performing an analysis. As noted above, ADaM datasets should be derived from the data contained in the SDTM datasets. There are features built into the ADaM standard that promote traceability from analysis results to ADaM datasets and from ADaM datasets to SDTM datasets. To ensure traceability, all SDTM variables utilized for variable derivations in ADaM should be included in the ADaM datasets when practical. Each submitted ADaM dataset should have its contents described with complete metadata in the define.xml file (See section 4.1.4.5) and within the ADRG as appropriate (See section 2.3).
符合 ADaM 的分析数据集的预期优势之一是,它们简化了执行分析所需的编程步骤。如上所述,ADaM 数据集应从 SDTM 数据集中包含的数据派生。ADaM 标准内置了一些功能,可促进从分析结果到 ADaM 数据集以及从 ADaM 数据集到 SDTM 数据集的可追溯性。为确保可追溯性,在可行的情况下,ADaM 中用于变量推导的所有 SDTM 变量都应包含在 ADaM 数据集中。每个提交的 ADaM 数据集都应在定义.xml文件(参见 4.1.4.5 节)和 ADRG(参见第 2.3 节)中描述其内容和完整的元数据。
4.1.2.3 Dataset Labels 数据集标签
Each dataset should be described by an internal label that is shown in the define.xml file. The label names of ADaM datasets should be different from those of the SDTM datasets. For example, the SDTM adverse event dataset (i.e., AE) and the ADaM adverse event dataset (i.e., ADAE) should not share the exact same dataset label, such as “Adverse Events.”
每个数据集都应由定义.xml文件中显示的内部标签进行描述。ADaM 数据集的标签名称应与 SDTM 数据集的标签名称不同。例如,SDTM 不良事件数据集(即 AE)和 ADaM 不良事件数据集(即 ADAE)不应共享完全相同的数据集标签,例如“不良事件”。
4.1.2.4 Subject-Level Analysis Data
Subject-Level Analysis Data (ADSL) is the subject-level analysis dataset for ADaM. All submissions containing standard analysis data should contain an ADSL file for each study. In addition to the variables specified for ADSL in the ADaMIG, such as those listed below in the core variables section (See section 4.1.2.5), the sponsor should include multiple additional variables representing various important baseline subject characteristics / covariates presented in the study protocol. Some examples of baseline characteristics / covariates for drug studies include, but are not limited to, disease severity scores such as Acute Physiology and Chronic Health Evaluation (APACHE) scores30, baseline organ function measurements such as calculated creatinine clearance or Forced Expiratory Volume in 1 second (FEV1), range categories for continuous variables, and numeric date variables in non-International Standards Organization (ISO) formats. Some examples of baseline characteristics for vaccine studies include, but are not limited to, past medical history (e.g., prior infection history), immunosuppressive conditions, prior vaccination history and concomitant medications/vaccines.
受试者级分析数据 (ADSL) 是 ADaM 的受试者级分析数据集。所有包含标准分析数据的提交都应包含每个研究的ADSL文件。除了在ADaMIG中为ADSL指定的变量,例如下面核心变量部分列出的变量(见第4.1.2.5节),申办者还应包括多个代表研究方案中提出的各种重要基线受试者特征/协变量的附加变量。药物研究的基线特征/协变量的一些示例包括但不限于疾病严重程度评分,例如急性生理学和慢性健康评估(APACHE)评分30,基线器官功能测量,例如计算的肌酐清除率或1秒内用力呼气量(FEV1),连续变量的范围类别以及非国际标准组织(ISO)格式的数字日期变量。疫苗研究基线特征的一些例子包括但不限于既往病史(例如,既往感染史)、免疫抑制性疾病、既往疫苗接种史和伴随药物/疫苗史。
4.1.2.5 Core Variables
Core variables, which include covariates presented in the study protocol that are necessary to analyze data, should be included in each ADaM dataset, and are typically already included in the ADSL dataset (See section 4.1.2.4). The core variables included in an ADaM dataset should be necessary for the analysis need in that dataset. Examples of core variables include study/protocol number, center/site number, geographic region, country, treatment assignment information, sex, age, race, analysis population flags (e.g., Intent-to-Treat (ITTFL), Full Analysis Set (FASFL), Safety (SAFFL), and Per-Protocol (PPROTFL)), and other important baseline demographic variables. Note that all variables that contain coded data should be accompanied by a variable that provides the decoded information.
核心变量,包括研究方案中提出的分析数据所必需的协变量,应包含在每个ADaM数据集中,并且通常已经包含在ADSL数据集中(参见第4.1.2.4节)。ADaM 数据集中包含的核心变量对于该数据集中的分析需求应该是必需的。核心变量的示例包括研究/方案编号、中心/站点编号、地理区域、国家/地区、治疗分配信息、性别、年龄、种族、分析人群标志(例如,意向治疗 (ITTFL)、完整分析集 (FASFL)、安全性 (SAFFL) 和按方案 (PPROTFL))以及其他重要的基线人口统计变量。请注意,所有包含编码数据的变量都应附带一个提供解码信息的变量。
In addition, it is important to note that SDTM datasets do not have core variables (such as demographic and population variables) repeated across the different domains. The duplication of core variables across various domains can be fulfilled through their inclusion in the corresponding analysis datasets. For example, the SDTM AE dataset does not allow for the inclusion of variables such as treatment arm, sex, age, or race. These and other variables should be included in the adverse event ADaM dataset (i.e., ADAE).
此外,请务必注意,SDTM 数据集没有跨不同域重复的核心变量(如人口统计和人口变量)。核心变量在各个域中的重复可以通过将其包含在相应的分析数据集中来实现。例如,SDTM AE 数据集不允许包含治疗手臂、性别、年龄或种族等变量。这些变量和其他变量应包含在不良事件 ADaM 数据集(即 ADAE)中。
4.1.2.6 Key Efficacy and Safety Data
Sponsors should submit ADaM datasets to support efficacy and safety analyses. At least one dataset should be referenced in the data definition file as containing the primary efficacy variables. Further, variables and parameters pertaining to the primary and secondary endpoints of a study, along with their derivations (as applicable), should be provided as well as documented appropriately (i.e., variable-level metadata or parameter value-level metadata) in the data definition file.
申办者应提交 ADaM 数据集以支持有效性和安全性分析。在数据定义文件中,应至少引用一个数据集作为包含主要有效性变量的数据集。此外,与研究的主要和次要终点有关的变量和参数及其推导(如适用)应提供并在数据定义文件中适当记录(即变量级元数据或参数值级元数据)。
4.1.2.7 Timing Variables
A variable for relative day of measurement or event, along with timing variables for visit, should be included when an ADaM dataset contains multiple records per subject (i.e., repeated measures data).
当 ADaM 数据集包含每个受试者的多个记录(即重复测量数据)时,应包括测量或事件的相对日期变量以及访问的时间变量。
4.1.2.8 Numeric Date Variables
Numeric date variables are needed for analysis and review purposes. Apply formats to all numeric date variables using a format that is understandable by SAS XPORT Version 5 files as per section 3.3.1 above. The software specific (as opposed to study specific) date of reference used to calculate numeric dates should be specified within the ADRG. In the event of partial dates, imputation should be performed only for dates required for analysis according to the SAP, and appropriate corresponding ADaM imputation flags should be utilized. When numeric time or date time variables are needed, all considerations apply as previously discussed for numeric dates.
分析和查看需要数字日期变量。根据上述 3.3.1 节,使用 SAS XPORT 版本 5 文件可以理解的格式,将格式应用于所有数字日期变量。用于计算数字日期的软件特定(而不是特定于研究)的参考日期应在ADRG中指定。如果是部分日期,应仅对根据 SAP 进行分析所需的日期执行插补,并应使用适当的相应 ADaM 插补标志。当需要数字时间或日期时间变量时,所有注意事项都适用,如前所述,适用于数字日期。
For traceability purposes, SDTM character dates formatted as ISO 8601 should also be included in the ADaM datasets.
出于可追溯性目的,格式化为 ISO 8601 的 SDTM 字符日期也应包含在 ADaM 数据集中。
4.1.2.9 Imputed Data 估算数据
When data imputation is utilized in ADaM, sponsors should submit the relevant supporting documentation (i.e., define.xml and ADRG) explaining the imputation methods.
当在ADaM中使用数据估算时,发起人应提交相关证明文件(即define.xml和ADRG),解释估算方法。
4.1.2.10 Software Programs
Sponsors should provide the software programs used to create all ADaM datasets and generate tables and figures associated with primary and secondary efficacy analyses. Furthermore, sponsors should submit software programs used to generate additional information included in Section 14 CLINICAL STUDIES of the Prescribing Information31, if applicable. The specific software utilized should be specified in the ADRG. Refer to FDA Statistical Software Clarifying Statement for more information. The main purpose of requesting the submission of these programs is to understand the process by which the variables for the respective analyses were created and to confirm the analysis algorithms and results. Sponsors should submit software programs in ASCII text format. Executable file extensions should not be used.
申办者应提供用于创建所有ADaM数据集的软件程序,并生成与主要和次要疗效分析相关的表格和数字。此外,申办者应提交用于生成其他信息的软件程序,这些信息包含在处方信息31的第14节临床研究中,如果适用。所使用的特定软件应在 ADRG 中指定。有关更多信息,请参阅 FDA 统计软件澄清声明。请求提交这些程序的主要目的是了解创建相应分析的变量的过程,并确认分析算法和结果。申办者应以 ASCII 文本格式提交软件程序。不应使用可执行文件扩展名 [译注:即程序文件后缀需改为.txt, 不能使用.sas]。
4.1.3 Standard for Exchange of Nonclinical Data 非临床数据交换标准
[译注:没有相关工作经验, 此章节略过]
4.1.4 General Considerations: SDTM, SEND, and/or ADaM 一般注意事项:SDTM、SEND 和/或 ADaM
4.1.4.1 Variables in SDTM and SEND: Required, Expected, and Permissible SDTM 和 SEND 中的变量:必需、预期和允许
For the purposes of SDTM and SEND submissions, all required, expected, and permissible variables that were collected, plus any variables that are used to compute derivations, should be submitted
出于 SDTM 和 SEND 提交的目的,应提交收集的所有必需、预期和允许变量,以及用于计算派生的任何变量。
FDA recognizes that SDTM contains certain operationally derived variables that have standard derivations across all studies (e.g., --STDY, EPOCH). If the data needed to derive these variables are missing, then these variables cannot be derived and the values should be null. The following are examples of some of the permissible and expected variables in SDTM and SEND that should be included, if available:
FDA认识到SDTM包含某些操作派生的变量,这些变量在所有研究中都有标准推导(例如,--STDY,EPOCH)。如果缺少派生这些变量所需的数据,则无法派生这些变量,并且值应为 null。以下是 SDTM 和 SEND 中应包含的一些允许变量和预期变量(如果可用)的示例:
1. Clinical baseline flags (e.g., last non-missing value prior to first dose) for laboratory results, vital signs, ECG, pharmacokinetic concentrations, and microbiology results. Nonclinical baseline flags (e.g., last non-missing value prior to first dose in parallel design studies) for laboratory results, vital signs, and ECG results. Currently for SDTM and SEND, baseline flags should be submitted if the data were collected or can be derived.
实验室结果、生命体征、心电图、药代动力学浓度和微生物学结果的临床基线标志(例如,首次给药前的最后一个非缺失值)。实验室结果、生命体征和心电图结果的非临床基线标志(例如,在平行设计研究中首次给药前的最后一个非缺失值)。目前,对于 SDTM 和 SEND,如果收集了数据或可以派生数据,则应提交基线标志。
2. EPOCH designators in SDTM. Please follow CDISC guidance for terminology. The variable EPOCH should be included for clinical subject-level observation (e.g., adverse events, laboratory, concomitant medications, exposure, and vital signs). This will allow the reviewer to easily determine during which phase of the study the observation occurred (e.g., screening, on-therapy, follow-up), as well as the actual intervention the subject experienced during that phase.
SDTM 中的 EPOCH 指示符。请遵循 CDISC 术语指南。对于临床受试者水平的观察(例如,不良事件、实验室、伴随药物、暴露和生命体征),应包括可变 EPOCH。这将使评价员能够轻松确定观察结果发生在研究的哪个阶段(例如,筛查,治疗,随访),以及受试者在该阶段经历的实际干预。
3. Whenever --DTC, --STDTC or --ENDTC, which have the role of timing variables, are included in a general observation class domain, the matching study day variables (--DY, --STDY, or --ENDY, respectively) should be submitted. For example, in most findings domains, --DTC is expected, which means that --DY should also be submitted. In the SDTM subject visits domain, SVSTDTC is required and SVENDTC is expected; therefore, both SVSTDY and SVENDY should be submitted.
每当具有时序变量作用的 --DTC、--STDTC 或 --ENDTC 包含在一般观测类域中时,应提交匹配的研究日变量(分别为 --DY、--STDY 或 --ENDY)。例如,在大多数发现域中,--DTC是预期的,这意味着--DY也应该提交。在SDTM受试者访问领域,SVSTDTC是必需的,SVENDTC是预期的;因此,应同时提交 SVSTDY 和 SVENDY。
As mentioned in section 4.1.3.3, in certain GLP nonclinical studies submitted in SEND, PCDTC and PCDY may be imputed.
如第4.1.3.3节所述,在SEND中提交的某些GLP非临床研究中,PCDTC和PCDY可能会被归因。
4.1.4.2 Dates in SDTM and SEND SDTM 和 SEND 域中的日期
Dates in SDTM and SEND domains should conform to the ISO 8601 format. Examples of how to implement dates are included in the SDTMIGs and SENDIGs
SDTM 和 SEND 域中的日期应符合 ISO 8601 格式。如何实现日期的示例包含在 SDTMIG 和 SENDIG 中
4.1.4.3 Naming Conventions in SDTM and SEND( SDTMIG 和 SENDIG 中指定的命名约定)
Naming conventions (variable name and label) and variable formats should be followed as specified in the SDTMIGs and SENDIGs.
应遵循 SDTMIG 和 SENDIG 中指定的命名约定(变量名称和标签)和变量格式。
4.1.4.4 SDTM and SEND Versions SDTM and SEND版本
When submitting clinical or nonclinical data, sponsors should not mix versions within a study. As noted above, the Catalog lists the versions that are supported by FDA.
在提交临床或非临床数据时,申办方不应在研究中混合使用。如上所述,该目录列出了FDA支持的版本。
4.1.4.5 Data Definition Files for SDTM, SEND, and ADaM (SDTM, SEND, and ADaM的数据说明文件)[译注:即define.xml]
The data definition file describes the metadata of the submitted electronic datasets, and is considered arguably the most important part of the electronic dataset submission for regulatory review. This data definition specification for submitted datasets defines the metadata structures that should be used to describe the datasets, variables, possible values of variables when appropriate, and controlled terminologies and codes. An insufficiently documented data definition file is a common deficiency that reviewers have noted. Consequently, the sponsor needs to provide complete detail in this file, especially for the specifications pertaining to derived variables. In addition, sponsors should also make certain that the code list and origin for each variable are clearly and easily accessible from the data definition file. The version of any external dictionary should be clearly stated both in the data definition file and in the full TS domain when it is submitted. The internal dataset label should also clearly describe the contents of the dataset. For example, the dataset label for an efficacy dataset might be ‘Time to Relapse (Efficacy).’
数据定义文件描述了所提交的电子数据集的元数据,并且可以说是提交电子数据集以供监管审查的最重要部分。此提交数据集的数据定义规范定义了应用于描述数据集、变量、变量的可能值(如果适用)以及受控术语和代码的元数据结构。记录不充分的数据定义文件是审稿人注意到的常见缺陷。因此,发起人需要在此文件中提供完整的详细信息,特别是对于与派生变量相关的规范。此外,发起人还应确保每个变量的代码列表和原点都可以从数据定义文件中清晰易行地访问。提交时,任何外部字典的版本都应在数据定义文件和完整的 TS 域中明确说明。内部数据集标签还应清楚地描述数据集的内容。例如,功效数据集的数据集标签可能是“复发时间(疗效)”。
Separate data definition files should be included for each type of electronic dataset submission, i.e., a separate data definition file for the SDTM datasets of a given clinical study, a separate data definition file for the SEND datasets of a given nonclinical study, and a separate data definition file for the ADaM datasets of a given clinical study. The data definition file should be submitted in XML format, i.e., a properly functioning define.xml. In addition to the define.xml, a printable define.pdf should be provided if the define.xml cannot be printed. To confirm that a define.xml is printable within the CDER IT environment, it is recommended that the sponsor submit a test version to cder-edata@fda.hhs.gov prior to application submission. The Catalog lists the currently supported version(s) of define.xml. It should be noted that define.xml version 2.0 is the preferred version. Sponsors should include a reference to the style sheet as defined in the specification (as listed in the Catalog) and place the corresponding style sheet in the same submission folder as the define.xml file. Within the eCTD study tagging file (STF), valid file-tags for define.xml are ‘data-tabulation-data-definition’ for SEND or SDTM datasets or ‘analysis-data-definition’ for ADaM datasets.
每种类型的电子数据集提交都应包含单独的数据定义文件,即给定临床研究的SDTM数据集的单独数据定义文件,给定非临床研究的SEND数据集的单独数据定义文件,以及给定临床研究的ADaM数据集的单独数据定义文件。数据定义文件应以XML格式提交,即功能正常的define.xml。除了define.xml之外,如果无法打印define.xml,还应提供可打印的define.pdf。为了确认define.xml在CDER IT环境中是可打印的,建议申办方在提交申请之前向 cder-edata@fda.hhs.gov 提交测试版本。目录列出了define.xml当前支持的版本。应该注意的是,定义.xml 2.0 版是首选版本。发起人应包括对规范中定义的样式表的引用(如目录中所列),并将相应的样式表放在与define.xml文件相同的提交文件夹中。在eCTD研究标记文件(STF)中,define.xml的有效文件标签是SEND或SDTM数据集的“数据制表数据定义”或ADaM数据集的“分析数据定义”。
4.1.4.6 Annotated Case Report Form (aCRF) for SDTM SDTM的带注释的病例报告表(aCRF)
An annotated case report form (aCRF) is a PDF document that maps the clinical data collection fields used to capture subject data (electronic or paper) to the corresponding variables or discrete variable values contained within the SDTM datasets. Regardless of whether the clinical database is in a format supported by the Catalog, an aCRF should be submitted preferably at the time a protocol is submitted. The aCRF should be provided as a PDF with the file name ‘acrf.pdf.’
带注释的病例报告表(aCRF)是一个PDF文档,用于将用于捕获受试者数据(电子或纸质)的临床数据收集字段映射到SDTM数据集中包含的相应变量或离散变量值。无论临床数据库是否采用目录支持的格式,都应在提交方案时提交aCRF。aCRF 应以 PDF 格式提供,文件名为“acrf.pdf”。
The aCRF should include treatment assignment forms, when applicable, and should map each variable on the CRF to the corresponding variables in the datasets (or database). The aCRF should include the variable names and coding for each CRF item.
aCRF应包括治疗分配表格(如果适用),并应将通用报告格式上的每个变量映射到数据集(或数据库)中的相应变量。aCRF应包括每个通用报告格式项目的变量名称和编码。
When data are recorded on the CRF but are not submitted, the CRF should be annotated with the text ‘NOT SUBMITTED.’ There should be an explanation in the relevant RG stating why these data have not been submitted.
如果数据记录在通用报告格式上但未提交,则应在通用报告格式上标注“未提交”的案文。在相关的RG中应该有一个解释,说明为什么这些数据没有被提交。
Section 5 Therapeutic Area Topics 治疗领域主题
[译注:未翻译,有需要时单独查阅]
Section 6 terminology 术语
6.1 General 综述
Common dictionaries should be used across all clinical studies and throughout the submission for each of the following: adverse events, concomitant medications, procedures, indications, study drug names, and medical history. FDA recommends that sponsors use, where appropriate, the terminologies supported and listed in the Catalog. It is important that coding standards, if they exist, be followed (e.g., ICH Medical Dictionary for Regulatory Activities (MedDRA) Term Selection: Points-to-Consider document). Frequently, sponsors submit data that do not conform to terminology standards, for example, misspelling of MedDRA or WHODrug Global terms, lack of conformance to upper / lower case, or the use of hyphens. All controlled terms submitted in datasets should conform to the exact case and spelling used by the terminology maintenance organization (e.g., MedDRA, CDISC controlled terminology). These conformance issues make it difficult to use or develop automated review and analysis tools. The use of a dictionary that is sponsor-defined or an extension of a standard dictionary should be avoided if possible, but, if essential, its use should be documented in the define.xml file and the relevant RGs.
通用词典应用于所有临床研究以及以下各项的整个提交过程中:不良事件,伴随药物,程序,适应症,研究药物名称和病史。FDA建议申办者在适当情况下使用目录中支持和列出的术语。重要的是要遵循编码标准(如果存在)(例如,ICH监管活动医学词典(MedDRA)术语选择:要考虑的要点文档)。通常,申办者提交的数据不符合术语标准,例如,MedDRA或WHODrug Global术语拼写错误,不符合大写/小写字母或使用连字符。数据集中提交的所有受控术语都应符合术语维护组织使用的确切大小写和拼写(例如,MedDRA、CDISC 控制术语)。这些一致性问题使得使用或开发自动审查和分析工具变得困难。如果可能,应避免使用由申办方人定义的字典或标准字典的扩展,但是,如果有必要,则应将其使用记录在define.xml文件和相关RG中。
6.1.1 Controlled Terminologies 受控术语
Controlled terminology standards are an important component of study data standardization and are a critical component of achieving semantically interoperable data exchange (See Appendix A). Generally, controlled terminology standards specify the key concepts that are represented as definitions, preferred terms, synonyms, codes, and code system.
受控术语标准是研究数据标准化的重要组成部分,是实现语义上可互操作的数据交换的关键组成部分(见附录A)。通常,受控术语标准指定表示为定义、首选术语、同义词、代码和代码系统的关键概念。
The analysis of study data is greatly facilitated by the use of controlled terms for clinical or scientific concepts that have standard, predefined meanings and representations. In electronic study data submissions, sponsors should provide the actual verbatim terms that were collected (e.g., on the CRF), as well as the coded term.
通过使用具有标准,预定义含义和表示的临床或科学概念的受控术语,极大地促进了研究数据的分析。在提交电子研究数据时,申办者应提供所收集的实际逐字逐句术语(例如,在通用报告格式上)以及编码术语。
Controlled terminology is also useful when consistently applied across studies to facilitate integrated analyses (that are stratified by study) and cross-study comparative analyses (e.g., when greater statistical power is needed to detect important safety signals). Cross-study comparisons and pooled integrated analyses occasionally provide critical information for regulatory decisions, such as statistical results that support effectiveness, as well as important information on exposure-response relationships and population pharmacokinetics
当在研究中一致地应用受控术语以促进综合分析(通过研究分层)和跨研究比较分析(例如,当需要更大的统计能力来检测重要的安全信号时)时,受控术语也是有用的。交叉研究比较和汇总综合分析偶尔会为监管决策提供关键信息,例如支持有效性的统计结果,以及有关暴露 - 反应关系和群体药代动力学的重要信息
6.1.2 Use of Controlled Terminologies 受控术语的使用
FDA recognizes that studies are conducted over many years, during which time versions of a terminology may change. Sponsors should use the most recent version of the dictionary available at the start of a clinical or nonclinical study. If a new version becomes available after the start of the study, sponsors may use the most current version of the dictionary for that clinical or nonclinical study. It is common to have different studies use different versions of the same dictionary within the same application (e.g., NDA, BLA). A submission of study data should describe (e.g., in the SDSP or relevant RG) the impact, if any, of the use of different versions on the study results. For example, if the sponsor anticipates pooling coded data across multiple studies, then it may be desirable to use a single version across those studies to facilitate pooling. If a sponsor selects this approach, then the approach and the justification should be documented in the Standardization Plan, or in an update to the plan.
FDA认识到研究是多年进行的,在此期间,术语的版本可能会发生变化。申办者应使用临床或非临床研究开始时可用的最新版本的词典。如果在研究开始后有新版本可用,申办者可以使用最新版本的字典进行临床或非临床研究。不同的研究在同一应用程序中使用同一字典的不同版本(例如,NDA,BLA)是比较常见的。提交的研究数据应描述(例如,在SDSP或相关RG中)使用不同版本对研究结果的影响(如果有的话)。例如,如果申办者预计在多个研究中汇集编码数据,那么在这些研究中使用单个版本以促进汇总可能是可取的。如果发起人选择此方法,则该方法和理由应记录在标准化计划中,或记录在计划的更新中。
Regardless of the specific versions used for individual studies, pooled analyses (e.g., for an ISS) should be conducted using a single version of a terminology. The current version should be used at the time that data across studies are pooled. This will ensure a consistent and coherent comparison of clinical and scientific concepts across multiple studies. Sponsors should specify the terminologies and versions used in the study in the relevant RG.
无论用于单个研究的具体版本如何,汇总分析(例如,ISS)应使用术语的单一版本进行。在合并各研究的数据时,应使用当前版本。这将确保在多项研究中对临床和科学概念进行一致和连贯的比较。申办者应在相关RG中指定研究中使用的术语和版本。
6.1.2.1 Use of the Specific Controlled Term ‘OTHER’ 使用特定受控术语“其他”
It is understood that the expansion of controlled terminology may lag behind scientific advancement, and that sometimes there may not be a relevant term within a controlled terminology’s value set to describe a clinical trial event, finding, or observation. However, it is not recommended to map a collected value to ‘OTHER’ when there is a controlled term available to match the collected value – even when the terminology allows for sponsor expansion. Each unique value in a --TERM field mapped to a --DECODE value of ‘OTHER’ should have a clear rationale outlined in the relevant RGs.
不言而喻,受控术语的扩展可能落后于科学进步,有时在受控术语的值集中可能没有相关术语来描述临床试验事件,发现或观察。但是,当有可控制的术语可用来匹配收集的值时,不建议将收集的值映射到“OTHER”,即使术语允许申办方扩展也是如此。映射到“OTHER”的 --DECODE 值的 --TERM 字段中的每个唯一值都应在相关 RG 中概述明确的理由。
6.1.3 Maintenance of Controlled Terminologies 维护受控术语
The use of supported controlled terminologies is recommended wherever available. If a sponsor identifies a concept for which no standard term exists, FDA recommends that the sponsor submit the concept to the appropriate terminology maintenance organization as early as possible to have a new term added to the standard dictionary. FDA considers this good terminology management practice. The creation of custom terms (i.e., so-called extensible code lists) for a submission is discouraged, because this does not support semantically interoperable study data exchange. Furthermore, the use of custom or extensible code lists should not be interpreted to mean that sponsors may substitute their own nonstandard terms in place of existing equivalent standardized terms. Sponsors should allow sufficient time for a proposed term to be reviewed and included in the terminology, as it is desirable to have the term incorporated into the standard terminology before the data are submitted. If custom terms cannot be avoided, the submitter should clearly identify and define them within the submission, reference them in the relevant RGs, and use them consistently throughout the application.
建议在可能的情况下使用受支持的受控术语。如果申办者确定一个不存在标准术语的概念,FDA建议申办者尽早将该概念提交给适当的术语维护组织,以便将新术语添加到标准词典中。FDA认为这是良好的术语管理实践。不鼓励为提交创建自定义术语(即所谓的可扩展代码列表),因为这不支持语义上可互操作的研究数据交换。此外,使用自定义或可扩展代码列表不应被解释为意味着发起人可以用自己的非标准术语代替现有的等效标准化术语。申办者应留出足够的时间对拟议的术语进行审查并纳入术语,因为在提交数据之前,最好将该术语纳入标准术语。如果无法避免自定义术语,提交者应在提交中清楚地标识和定义它们,在相关RG中引用它们,并在整个应用程序中一致地使用它们。
If a sponsor identifies an entire information domain for which FDA has not accepted a specific standard terminology, the sponsor may select a standard terminology to use, if one exists. FDA recommends that sponsors include this selection in the Standardization Plan (See section 2.1) or in an update to the existing plan, and reference it in the relevant RG. If no controlled terminology exists, the sponsor may define custom terms. For clinical studies, the non-FDA supported terms (whether from a non-supported standard terminology or sponsor-defined custom terms) should then be used consistently throughout all relevant studies within the application. Although the consistent use of non-FDA supported terms across all nonclinical studies within an application is recommended, it is understood that that this may not always be possible.
如果申办者确定了FDA尚未接受特定标准术语的整个信息域,则申办者可以选择要使用的标准术语(如果存在)。FDA建议申办者将此选择纳入标准化计划(见第2.1节)或现有计划的更新中,并在相关RG中引用。如果不存在受控术语,发起人可以定义自定义术语。对于临床研究,非FDA支持的术语(无论是来自不支持的标准术语还是申办者定义的自定义术语)应在申请内的所有相关研究中一致使用。尽管建议在申请中的所有非临床研究中一致使用非FDA支持的术语,但据了解,这可能并不总是可能的。
6.2 CDISC Controlled Terminology CDISC控制术语
Sponsors should use the terminologies and code lists in the CDISC Controlled Terminology, which can be found at the NCI (National Cancer Institute) Enterprise Vocabulary Services.52 For variables for which no standard terms exists, or if the available terminology is insufficient, the sponsor should propose its own terms. The sponsor should provide this information in the define.xml file and in the relevant RGs.
申办者应使用CDISC管制术语中的术语和代码列表,这些术语和代码列表可在NCI(国家癌症研究所)企业词汇服务中找到。对于不存在标准术语的变量,或者如果可用术语不足,申办者应提出自己的术语。发起人应在define.xml文件和相关RG中提供此信息。
6.3 Adverse Events
6.3.1 MedDRA
6.3.1.1 General Considerations 一般注意事项
MedDRA is used for coding adverse events.Generally, the studies included in an application are conducted over many years and may have used different MedDRA versions. The expectation is that sponsors or applicants will use the most current version of MedDRA at the time of study start. However, there is no requirement to recode earlier studies
MedDRA用于编码不良事件3通常,申请中包含的研究是多年进行的,并且可能使用了不同的MedDRA版本。期望申办者或申请人将在研究开始时使用最新版本的MedDRA。但是,不需要对早期研究进行重新编码
The spelling and capitalization of MedDRA terms should match the way the terms are presented in the MedDRA dictionary (e.g., spelling and case). Common errors that have been observed include the incorrect spelling of a System Organ Class (SOC) and other MedDRA terms.
MedDRA 术语的拼写和大小写应与 MedDRA 字典中术语的显示方式相匹配(例如,拼写和大小写)。观察到的常见错误包括系统器官类 (SOC) 和其他 MedDRA 术语的拼写错误。
To avoid potential confusion or incorrect results, the preparation of the adverse event dataset for the ISS should include MedDRA terms from the most current version of MedDRA at the time that data across studies are pooled. The reason for an ISS based on a single version of MedDRA is that reviewers often analyze adverse events across studies, including the use of Standardized MedDRA Queries. In addition, sponsors should use the MedDRA-specified hierarchy of terms. The SDTM variables for the different hierarchy levels should represent MedDRA-specified primary SOC-coded terms.
为了避免潜在的混淆或不正确的结果,ISS不良事件数据集的准备应包括合并不同研究数据时最新版本的MedDRA术语。基于单一版本的MedDRA的国际空间站的原因是,评价员经常分析研究中的不良事件,包括使用标准化MedDRA查询.此外,申办者应使用MedDRA指定的术语层次结构。不同层次结构级别的 SDTM 变量应表示 MedDRA 指定的主要 SOC 编码术语。
6.4 Medications
6.4.1 FDA Unique Ingredient Identifier FDA唯一成分标识符
6.4.1.1 General Considerations 一般注意事项
The Unique Ingredient Identifier (UNII) should be used to identify active ingredients (specifically, active moieties) that are administered to investigational subjects in a study (either clinical or nonclinical). This information should be provided in the SDTM TS domain. UNIIs should be included for all active moieties of investigational products (TSPARMCD= TRT or TRTUNII), active comparators (TSPARMCD= COMPTRT), and any protocol-specified background treatments (TSPARMCD= CURTRT).
应使用唯一成分标识符(UNII)来识别在研究(临床或非临床)中给予研究对象的活性成分(特别是活性部分)。此信息应在 SDTM TS 域中提供。对于研究性产品(TSPARMCD= TRT 或 TRTUNII)、有源比较器 (TSPARMCD = COMPTRT) 和任何方案指定的背景处理 (TSPARMCD = CURTRT) 的所有活性部分,都应包括 UNII。
If a medicinal product has more than one active moiety, then multiple records in the full TS should be provided, one for each active moiety. For example, if the investigational product is Bactrim (a combination of sulfamethoxazole and trimethoprim), then TS will contain two records for TSPARMCD= TRT: one for sulfamethoxazole and one for trimethoprim.
如果医药产品具有多个活性部分,则应在完整的TS中提供多个记录,每个活性部分一个记录。例如,如果研究产品是Bactrim(磺胺甲噁唑和甲氧苄啶的组合),则TS将包含TSPARMCD= TRT的两条记录:一条用于磺胺甲噁唑,一条用于甲氧苄啶。
The preferred substance names and UNII codes can be found by searching FDA’s Substance Registration System, hosted by the National Library of Medicine. We recognize that unapproved substances may not yet have registered UNII codes. We recommend that sponsors obtain UNII codes for unapproved substances as early in drug development as possible, so that relevant information, such as study data, can be unambiguously linked to those substances.
通过搜索由国家医学图书馆托管的FDA物质注册系统可以找到首选的物质名称和UNII代码.我们认识到未经批准的物质可能尚未注册UNII代码。我们建议申办者在药物开发过程中尽早获得未经批准的物质的UNII代码,以便相关信息(如研究数据)可以明确地与这些物质联系起来
6.4.2 WHODrug Global
6.4.2.1 General Considerations 一般注意事项
World Health Organization (WHO) Drug Global57 is a dictionary maintained and updated by Uppsala Monitoring Centre. WHODrug Global contains unique product codes for identifying drug names and listing medicinal product information, including active ingredients and therapeutic uses.
世界卫生组织(WHO) Drug Global是由Uppsala 监测中心维护和更新的词典。WHODrug Global包含用于识别药物名称和列出药品信息(包括活性成分和治疗用途)的唯一产品代码。
Typically, WHODrug Global is used to code concomitant medications. The variable --DECOD should be populated with the active substances from the WHODrug Global Dictionary, and --CLAS populated with the drug class.
通常,WHO全球药物用于对伴随的药物进行编码。变量 --DECOD 应填充 WHODrug 全球词典中的活性物质,并使用 --CLAS 填充药物类别。
When using WHODrug Global, --CLAS is recommended to be populated with the Anatomic Therapeutic Chemical (ATC) class most suitable per intended use, and the remainder of the ATC classes, if any, placed in SUPPCM. Alternately, the use of the SUPPCM or FACM domains to populate all ATC Classes associated with the --DECOD value is acceptable. ATC classes should be submitted at the fourth level or most specific available as defined within WHODrug Global.
当使用WHODrug Global时,建议使用最适合每种预期用途的解剖治疗化学品(ATC)类,并将ATC类的其余部分(如果有的话)放在SUPPCM中。或者,使用 SUPPCM 或 FACM 域来填充与 --DECOD 值关联的所有 ATC 类是可以接受的。ATC课程应在第四级或世卫组织全球药物中定义的最具体级别提交。
Generally, studies included in a submission are conducted over many years and may have used different WHODrug Global versions to code concomitant medications. The expectation is the most current B3-format annual version of WHODrug Global at the time of study start will be used to code concomitant medications. There is no requirement to recode earlier studies to align with the WHODrug Global version of later studies.
一般来说,提交材料中包含的研究是多年进行的,并且可能使用不同的WHODrug Global版本来编码伴随的药物。预计在研究开始时,最新的B3格式的WHODrug Global年度版本将用于编码伴随的药物。不需要对早期研究进行重新编码,以与后来的WHODrug全球版本保持一致。
6.5 Pharmacologic Class 药理学类
6.5.1 Medication Reference Terminology 药物参考术语
6.5.1.1 General Considerations 一般注意事项
The Veterans Administration’s Medication Reference Terminology (MED-RT)58 should be used to identify the pharmacologic class(es) of all active investigational substances that are used in a study (either clinical or nonclinical). This information should be provided in the SDTM TS domain when a full TS is indicated. The information should be provided as one or more records in TS, where TSPARMCD= PCLAS.
退伍军人管理局的药物参考术语(MED-RT)应用于识别研究中使用的所有活性研究物质(临床或非临床)的药理学类别。当指示完整的 TS 时,应在 SDTM TS 域中提供此信息。该信息应作为 TS 中的一条或多条记录提供,其中 TSPARMCD = PCLAS。
Pharmacologic class is a complex concept that is made up of one or more component concepts: mechanism of action (MOA), physiologic effect (PE), and chemical structure (CS).59 The established pharmacologic class is generally the MOA, PE, or CS term that is considered the most scientifically valid and clinically meaningful. Sponsors should include in TS (the full TS) the established pharmacologic class of all active moieties of investigational products used in a study. FDA maintains a list of established pharmacologic classes of approved moieties.60 If the established pharmacologic class is not available for an active moiety, then the sponsor should discuss the appropriate MOA, PE, and CS terms with the review division. For unapproved investigational active moieties where the pharmacologic class is unknown, the PCLAS record may not be available.
药理学类是一个复杂的概念,由一个或多个组分概念组成:作用机制(MOA),生理效应(PE)和化学结构(CS)。已建立的药理学类别通常是MOA,PE或CS术语,被认为是最科学有效和临床上最有意义的。申办者应在 TS(完整 TS)中包括研究中使用的所有活性部分的既定药理学类别。FDA维护已批准的部分的已建立药理学类别的列表。如果已建立的药理学类别不适用于活性部分,则申办者应与评价部门讨论适当的MOA,PE和CS术语。对于药理学类别未知的未经批准的研究活性部分,PCLAS记录可能不可用。
6.6 Indication 适应症
6.6.1 SNOMED CT
6.6.1.1 General Considerations 一般注意事项
The International Health Terminology Standards Organization’s (IHTSDO) Systematized Nomenclature of Medicine – Clinical Terms (SNOMED CT)61 should be used to identify the medical condition or problem that the investigational product in a study is intended to affect (treat, diagnose or prevent, i.e., the indication). This information should be provided in the SDTM TS domain (the full TS) as a record where TSPARMCD= INDIC and TSPARMCD= TDIGRP. SNOMED CT was chosen to harmonize with Indication information in Structured Product Labeling (SPL). Because the granted indication may include important qualifiers to fulfill the need for adequate directions for use (e.g., descriptors of the population to be treated, adjunctive or concomitant therapy, or specific tests needed for patient selection), the indication section in a label may not be fully represented by available SNOMED CT codes.
国际卫生术语标准组织(IHTSDO)的医学系统化命名法– 临床术语(SNOMED CT)应用于确定研究中的研究产品旨在影响的医疗状况或问题(治疗,诊断或预防,即适应症)。此信息应在 SDTM TS 域(完整的 TS)中作为记录提供,其中 TSPARMCD = INDIC 和 TSPARMCD = TDIGRP。选择SNOMED CT是为了与结构性产品标签(SPL)中的适应症信息相协调。由于授予的适应症可能包括重要的限定词,以满足对适当使用方向的需求(例如,待治疗人群的描述符,辅助或伴随治疗,或患者选择所需的特定测试),因此标签中的适应症部分可能没有完全由可用的SNOMED CT代码表示。
6.7 Laboratory Tests 实验室检查
6.7.1 LOINC
6.7.1.1 General Considerations 一般注意事项
The Logical Observation Identifiers Names and Codes (LOINC) is a clinical terminology housed by the Regenstrief Institute LOINC codes are universal identifiers for laboratory and other clinical observations that enable semantically interoperable clinical data exchange. The laboratory portion of the LOINC database contains the categories of chemistry, hematology, serology, microbiology (including parasitology and virology), toxicology, and more. The SDTM standard supports LOINC codes using the LBLOINC variable. LOINC codes should not be added to SEND datasets.
逻辑观测标识符 名称和代码(LOINC)是Regnestrief Institute的临床术语LOINC代码是实验室和其他临床观察的通用标识符,可实现语义上可互操作的临床数据交换。LOINC数据库的实验室部分包含化学,血液学,血清学,微生物学(包括寄生虫学和病毒学),毒理学等类别。SDTM 标准支持使用 LBLOINC 变量的 LOINC 代码。不应将 LOINC 代码添加到 SEND 数据集
When submitting LOINC codes you should:
1) Continue submitting laboratory data in the CDISC SDTM format using CDISC laboratory terminology alongside the LOINC code for a given laboratory test.
2) Enter LOINC codes in the LBLOINC field of the SDTM LB domain and populate LBMETHOD when available. When LOINC codes are unavailable, leave the field blank.
3) Submit LOINC codes only when they are available from the clinical laboratories as a pass-through only, i.e. reporting the codes as received from the laboratories with no modifications. FDA understands that there may be inconsistencies in the specification and interpretation of LOINC codes submitted across tests, studies, and subjects.
4) Provide in-vitro diagnostic (IVD) device information in the SDTM Device Identifiers (DI) domain, when available. This information will help inform further FDA guidance on the consistency of LOINC codes associated with laboratory devices.
提交LOINC代码时,您应该:
1) 继续以 CDISC SDTM 格式提交实验室数据,使用 CDISC 实验室术语以及给定实验室测试的 LOINC 代码。
2) 在 SDTM LB 域的 LBLOINC 字段中输入 LOINC 代码,并在可用时填充 LBMETHOD。当 LOINC 代码不可用时,请将该字段留空。
3) 仅当 LOINC 代码仅作为直通从临床实验室获得时提交 LOINC 代码,即报告从实验室收到的代码,无需修改。FDA了解,在测试,研究和受试者之间提交的LOINC代码的规范和解释可能存在不一致之处。
4)在SDTM设备标识符(DI)域中提供体外诊断(IVD)设备信息(如果可用)。这些信息将有助于为FDA关于实验室设备相关LOINC代码一致性的进一步指导提供信息。
7. Electronic Submission Format 电子提交格式
7.1 eCTD Specifications eCTD规范
For information on how to incorporate datasets into the eCTD, please reference the Guidance to Industry Providing Regulatory Submissions in Electronic Format: Certain Human Pharmaceutical Product Applications and Related Submissions Using the Electronic Common Technical Document Specifications. Information on eCTD Validations, including those referenced in the “Technical Rejection Criteria for Study Data Important Information” (Appendix F), can be found in the Specifications for eCTD Validation Criteria. Details on the expectations for validations applying to study data can be found in Section 8.2.2 (Support on Data Validation Rules) and Appendix F (Technical Rejection Criteria for Study Data Important Information) of this Technical Conformance Guide.
有关如何将数据集纳入eCTD的信息,请参阅《以电子格式提供监管提交的行业指南:某些人类医药产品应用和使用电子通用技术文件规范的相关提交内容》。 关于eCTD验证的信息,包括“研究数据重要信息的技术拒绝标准”(附录F)中引用的那些信息, 可以在eCTD验证标准的规范中找到。有关适用于研究数据的验证期望的详细信息,请参阅本技术一致性指南的第 8.2.2 节(数据验证规则支持)和附录 F(研究数据重要信息的技术拒绝标准)。
The study identifier (STUDYID) in trial summary (TS) and [study-id] in the study tagging file (STF)) should be identical wherever possible. For studies where alignment of the study identifier across TS and STF is not feasible, the value for [study-id] used in the STF should be included in TS using the parameter SPREFID. Though SPREFID is not in the SDTM controlled terminology for TSPARMCD, please use SPREFID to reconcile study identifiers where necessary for SEND or SDTM studies. FDA will use SPREFID to match study identifiers across STF and TS to establish the study start date where necessary for evaluation against the eCTD validation criteria.
试验摘要 (TS) 中的研究标识符 (STUDYID) 和研究标记文件 (STF) 中的 [study-id] 应尽可能相同。对于在 TS 和 STF 之间无法对齐研究标识符的研究,STF 中使用的 [study-id] 的值应使用参数 SPREFID 包含在 TS 中。虽然SPREFID不在TSPARMCD的SDTM受控术语中,但在SEND或SDTM研究必要时使用SPREFID来协调研究标识符。FDA将使用SPREFID来匹配STF和TS的研究标识符,以便在必要时根据eCTD验证标准进行评估以确定研究开始日期。
Do not use the eCTD ‘append’ lifecycle operator when submitting updated or changed content within study data files that were previously submitted. Updated files should be submitted using the ‘replace’ operator.
在提交以前提交的研究数据文件中更新或更改的内容时,请勿使用 eCTD“追加”生命周期运算符。应使用“替换”运算符提交更新的文件。
When nonclinical Weight of Evidence (WOE) documents are submitted to the Agency as assessments for particular topics or as justification of why a toxicity study is not needed, it is recommended that these toxicity risk assessments are submitted to the nonclinical eCTD Modules relevant to the topic. Examples are listed below:
Rodent Carcinogenicity: Module 4.2.3.4
Reproductive and Developmental Toxicity: Module 4.2.3.5
Juvenile Animal Toxicity: Module 4.2.3.5
当非临床证据权重(WOE)文件作为特定主题的评估或作为为什么不需要毒性研究的理由提交给该机构时,建议将这些毒性风险评估提交给与该主题相关的非临床eCTD模块。示例如下:
啮齿动物致癌性:模块4.2.3.4
生殖和发育毒性:模块4.2.3.5
幼年动物毒性:模块4.2.3.5
Cross-reference to these WOE documents may also be included within eCTD Module 2.4 (Nonclinical Overview summaries). Supporting literature references submitted with any WOE document should be submitted to eCTD Module 4.3 (Literature References). When a WOE document is submitted to an eCTD module that is subject to the Technical Rejection Criteria (e.g., carcinogenicity risk assessment submitted to Module 4.2.3.4), a simplified ts.xpt file must accompany this document. The TSVALNF field of the simplified ts.xpt file should be populated with the null value “NA” (Not Applicable) as further described under Section 8.2.2 (Support on Data Validation Rules) of this Technical Conformance Guide.
对这些WOE文件的交叉引用也可以包含在eCTD模块2.4(非临床概述摘要)中。与任何WOE文件一起提交的支持文献参考文献应提交给eCTD模块4.3(文献参考文献)。当 WOE 文档提交到受技术拒绝标准约束的 eCTD 模块(例如,提交到模块 4.2.3.4 的致癌性风险评估)时,本文档必须随附一个简化的 ts.xpt 文件。简化的 ts.xpt 文件的 TSVALNF 字段应填充空值“NA”(不适用),如本技术一致性指南的第 8.2.2 节(数据验证规则支持)中所述。
7.2 Electronic File Directory 电子文件目录
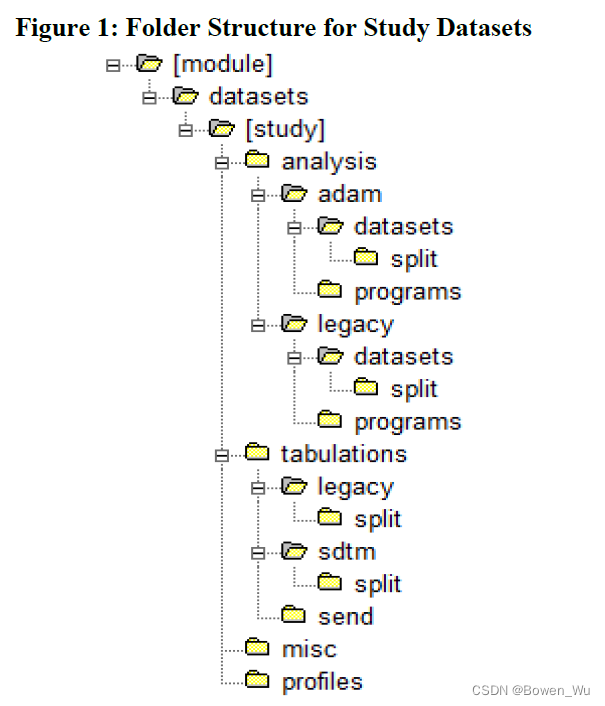
Study datasets and their supportive files should be organized into a specific file directory structure when submitted in the eCTD format (See Figure 1 and Table 2 below). Note that this structure is distinct from the eCTD headings and hierarchy folder structure, and does not affect it. Submission of files within the appropriate folders allows automated systems to detect and prepare datasets for review, and minimizes the need for manual processing.
以eCTD格式提交时,研究数据集及其支持文件应组织成特定的文件目录结构(见下面的图1和表2)。请注意,此结构与 eCTD 标题和层次结构文件夹结构不同,不会影响它。在适当的文件夹中提交文件允许自动化系统检测和准备数据集以供查看,并最大限度地减少手动处理的需求。
If you need to split a file that exceeds file size limits (See section 3.3.2), you should submit the smaller split files in the ‘split’ sub-folder in addition to the larger non-split file in the original data folder. There is no need for a second define.xml file to be submitted within the split subfolder.
如果需要拆分超过文件大小限制的文件(请参阅第 3.3.2 节),除了提交原始数据文件夹中较大的非拆分文件外,还应在“split”子文件夹中提交较小的拆分文件。无需在拆分子文件夹中提交第二个define.xml文件。
For rodent carcinogenicity studies submitted in 4.2.3.4, the tumor.xpt file and its associated define.pdf should be placed in analysis\legacy\datasets subfolder under the study datasets folder.
对于在4.2.3.4中提交的啮齿动物致癌性研究,tumor.xpt文件及其相关define.pdf应放置在研究数据集文件夹下的analyment\legacy\datasets子文件夹中。
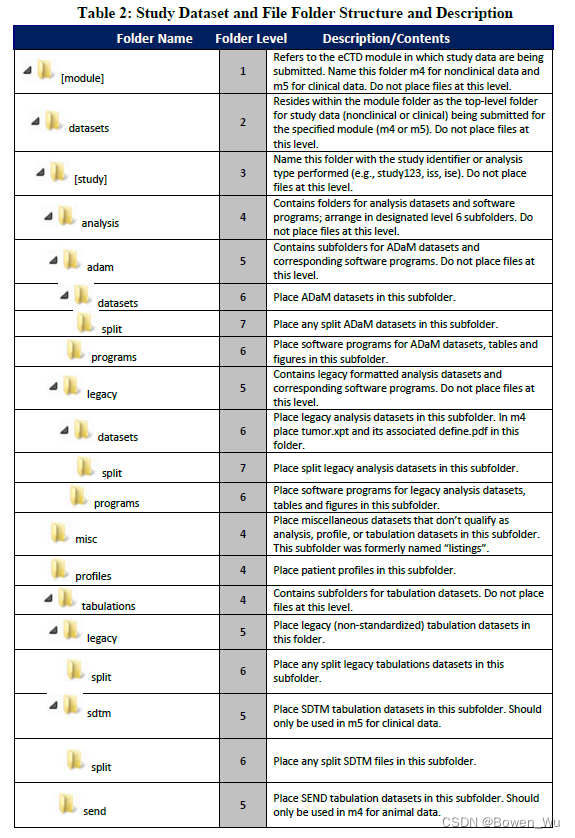
The file folder structure for study datasets is summarized in Figure 1. Table 2 provides the study dataset and file folder structure and associated description. For more detailed examples of file folder structures for clinical and non-clinical datasets in both standardized and legacy formats, please see Appendix E: Example Study Data Folder Structure.
图 1 总结了研究数据集的文件夹结构。表 2 提供了研究数据集和文件夹结构以及相关的描述。有关标准化和传统格式的临床和非临床数据集的文件夹结构的更多详细示例,请参阅附录 E:示例研究数据文件夹结构。
7.3 eCTD Sample Submission eCTD样品提交
The FDA would like to work closely with people who plan to provide a submission using the eCTD specifications and offer to help smooth the process. The Agency also offers a process for submitting sample standardized datasets for validation. Sample submissions are tests only and not considered official submissions. They are not reviewed by FDA reviewers at any time. The Electronic Submissions page provides more information regarding the test submission process.
FDA希望与计划使用eCTD规范提供提交并帮助顺利完成流程的人员密切合作。机构还提供了提交标准化数据集样本进行验证的流程。样本提交只是测试,不被视为正式提交。FDA审查员在任何时候都不会对其进行审查。“电子提交”页面提供了有关测试提交过程的更多信息。
8. Study Data Validation and Traceability 研究数据验证和可追溯性
8.1 Definition of Study Data Validation 研究数据验证的定义
Study data validation helps to ensure that the study data are compliant, useful, and will support meaningful review and analysis. Validation activities occur at different times during submission and review of study data, including submission receipt and at the beginning of the regulatory review. Validation of study data that occurs upon receipt of a submission follows the process for Technical Rejection Criteria for Study Data (See Appendix F).
研究数据验证有助于确保研究数据合规、有用,并支持有意义的审查和分析。验证活动在提交和审查研究数据期间的不同时间进行,包括提交收到和监管审查开始时。在收到提交后对研究数据进行验证遵循研究数据技术拒绝标准的过程(见附录F)。
8.2 Types of Study Data Validation Rules 研究数据类型 数据验证规则
1. Standards Development Organizations (e.g., CDISC) provide rules that assess conformance to its published standards (See www.CDISC.org).
2. FDA eCTD Technical Rejection Criteria for Study Data that assess conformance to the standards listed in the Catalog (See section 7.1, section 8.2.2, and Appendix F).
3. FDA Business and Validator rules to assess that the data support regulatory review and analysis.
1.标准制定组织(例如,CDISC)提供评估是否符合其已发布标准的规则(见 www.CDISC.org)。
2. FDA eCTD技术拒绝标准的研究数据,用于评估是否符合目录中列出的标准(见第7.1节,第8.2.2节和附录F)。
3. FDA业务和验证器规则,用于评估数据是否支持监管审查和分析。
8.2.1 FDA Business and Validator Rules FDA业务和验证规则
FDA Business Rules describe the business requirements for regulatory review to help ensure that study data are compliant and useful and support meaningful review and analysis. The list of business rules will grow and change with experience and cross-center collaborations. All business rules should be followed where applicable. The business rules are accompanied with validator rules which provide details regarding FDA's assessment of study data for purposes of review and analysis. The FDA Validator Rules also represent the latest understanding of what best supports regulatory review. The Study Data Standards Resources webpage page provides links to the currently available FDA Business and Validator rules.
FDA业务规则描述了监管审查的业务需求,以帮助确保研究数据合规和有用,并支持有意义的审查和分析。业务规则列表将随着经验和跨中心协作而增长和变化。在适用的情况下,应遵循所有业务规则。业务规则附有验证器规则,这些规则提供有关FDA评估研究数据的详细信息,以便进行审查和分析。FDA验证规则也代表了对最能支持监管审查的最新理解。研究数据标准资源网页页面提供了当前可用的FDA业务和验证器规则的链接。
8.2.2 Support on Data Validation Rules 对数据验证规则的支持
Sponsors should evaluate their study data before submission against the conformance rules published by an SDO, the eCTD Technical Rejection Criteria for Study Data (See Appendix F), and the FDA Business Rules. Sponsors may also wish to use the FDA Validator Rules to understand what is available to the FDA reviewer. Sponsors should either correct any discrepancies between study data and the standard or the business rules or explain meaningful discrepancies in the relevant Reviewer Guide (RG). Additional information about conformance to the standard, FDA Business Rules, or FDA Validator Rules that could facilitate review of the submitted data, or establish consistency and traceability between the study data and the Study Report, should also be provided in the relevant RG.
申办者应在提交之前根据SDO发布的一致性规则,eCTD研究数据的技术拒绝标准(见附录F)和FDA业务规则评估其研究数据。申办者还可能希望使用FDA验证规则来了解FDA审查员可以获得什么。申办者应纠正研究数据与标准或业务规则之间的任何差异,或在相关审稿人指南(RG)中解释有意义的差异。有关是否符合标准,FDA业务规则或FDA验证者规则的其他信息,这些信息可能有助于审查提交的数据,或在研究数据和研究报告之间建立一致性和可追溯性,也应在相关的RG中提供。
8.2.2.1 eCTD Technical Rejection Criteria for Study Data (See Appendix F for more details) eCTD研究数据技术拒绝标准(详见附录F)
FDA implemented an approach to determine compliance with the requirement to submit electronic standardized study data. The technical rejection criteria are automated validations by the Center (CDER or CBER) inbound processing system using the FDA Specifications for eCTD Validation Criteria68 as described below.
FDA实施了一种方法来确定是否符合提交电子标准化研究数据的要求。技术拒绝标准是中心(CDER或CBER)入站处理系统使用Specifications for eCTD Validation Criteria进行的自动验证,如下所述
In order for the FDA automated eCTD validation process to determine the study start date (SSD) for the submitted study, FDA relies on the SSD value provided in the Trial Summary dataset (ts.xpt) that is referenced in the Study Tagging File (STF).69 This validation confirms the submission of a valid STF (eCTD validation error 1789) and a Trial Summary (TS) domain (eCTD validation error 1734). For a nonclinical study that contains a study report with file tags “pre-clinical-study-report,” “legacy-clinical-study-report,” or “study-report-body,” and/or an xpt formatted dataset, the expectation for content in the TS domain (simplified or full)70 depends on whether the study is submitted in compliance with a CDISC standard. Appendix G (Examples of ts.xpt Datasets) provides the appropriate content and an example of the TS domains for each case. The expectation is that when the SSD is after the established required deadlines, the study data must comply with the standards in the FDA Data Standards Catalog. The validation will then identify that the required dataset files (eCTD validation error 1736) are under the correct file tag within the STF (eCTD validation error 1735). If there are no high validation errors within the eCTD submission, the submission will continue to be processed.
为了使 FDA 自动化 eCTD 验证流程确定所提交研究的研究开始日期 (SSD),FDA 依赖于研究标记文件 (STF) 中引用的试验摘要数据集 (ts.xpt) 中提供的 SSD 值。69 此验证确认提交了有效的 STF(eCTD 验证错误 1789)和试验摘要 (TS) 域(eCTD 验证错误 1734)。对于包含文件标签为“临床前研究报告”、“遗留临床研究报告”或“研究报告正文”的研究报告和/或 xpt 格式数据集的研究报告的非临床研究报告,对 TS 域(简化或完整)70 中内容的期望取决于提交的研究是否符合 CDISC 标准。附录 G(ts.xpt 数据集示例)提供了适用于每种情况的相应内容和 TS 域示例。预期是,当SSD在规定的所需截止日期之后时,研究数据必须符合FDA数据标准目录中的标准。然后,验证将确定所需的数据集文件(eCTD 验证错误 1736)位于 STF 中的正确文件标记下(eCTD 验证错误 1735)。如果 eCTD 提交中没有高验证错误,则将继续处理提交。
8.2.2.2 Technical Rejection Criteria and Use of a Simplified ts.xpt for Clinical Studies 技术拒绝标准和临床研究简化ts.xpt的使用
Technical rejection criteria have been added to the Specifications for eCTD Validation Criteria to determine compliance with the requirements for submitting standardized study data7when xpt formatted datasets are submitted to FDA in TRC applicable sections within Module 5.
技术拒绝标准已添加到eCTD验证标准规范中,以确定是否符合提交标准化研究数据的要求当xpt格式的数据集在模块5的TRC适用部分中提交给FDA时。
When a xpt formatted dataset is submitted, the STF for the study is then checked for the presence of a trial summary (TS) file (full or simplified). A full ts.xpt file would be expected when the study type and study initiation date meet the criteria for requiring SDTM and ADaM datasets as described in the current FDA Data Standards Catalog.
提交 xpt 格式的数据集时,将检查研究的 STF 是否存在试验摘要 (TS) 文件(完整或简化)。当研究类型和研究开始日期满足当前FDA数据标准目录中所述的SDTM和ADaM数据集要求的标准时,将需要完整的ts.xpt文件。
There are cases in which a xpt formatted dataset submitted to TRC applicable sections within eCTD Module 5 using one of the STFs (see sections 7.1, 8.2.2.1, and Appendix F) is not required to include accompanying SDTM and ADaM datasets. In such cases, a simplified ts.xpt file should be included with the xpt formatted dataset. A simplified ts.xpt file serves to provide limited machine-readable information such that any submitted xpt formatted dataset not requiring SDTM and ADaM datasets will be appropriately identified by the Center’s processing system.
在某些情况下,使用其中一个STF(参见第7.1节,8.2.2.1节和附录F)提交给eCTD模块5中TRC适用部分的xpt格式数据集不需要包括随附的SDTM和ADaM数据集。在这种情况下,简化的 ts.xpt 文件应包含在 xpt 格式的数据集中。简化的ts.xpt文件用于提供有限的机器可读信息,以便中心处理系统将适当地识别任何不需要SDTM和ADaM数据集的提交的xpt格式数据集。
也可能存在SDTM和ADaM不需要的情况,即使该研究在2016年12月17日之后开始。以下列表包含可能的示例(不是详尽无遗的列表):
•提交给ANDA申请的试点研究
•提交给ANDA申请的失败研究
如果 SDTM 和 ADaM 在 2016 年 12 月 17 日之后开始的研究不适用,请参阅附录 G,示例 B 了解应使用的简化 ts.xpt 文件的格式,其中 TSVALNF 字段将使用空值“NA”(不适用)进行填充。
8.2.2.3 Technical Rejection Criteria and Use of a Simplified ts.xpt for Nonclinical Studies (eCTD Modules 4.2.3.1, 4.2.3.2, and 4.2.3.4) for CDER CDER的技术拒绝标准和简化ts.xpt在非临床研究中的应用(eCTD模块4.2.3.1,4.2.3.2和4.2.3.4)
[译注:略]
8.3 Study Data Traceability 研究数据可追溯性
8.3.1 Overview 概述
An important component of a regulatory review is an understanding of the provenance of the data (e.g., traceability of the sponsor’s results back to the CRF data). Traceability permits an understanding of the relationships between the analysis results (tables, listings and figures in the study report), analysis datasets, tabulation datasets, and source data. Traceability enables the reviewer to accomplish the following:
监管审查的一个重要组成部分是对数据来源的理解(例如,申办者的结果可追溯到通用报告格式数据)。可追溯性允许了解分析结果(研究报告中的表格、列表和数字)、分析数据集、制表数据集和源数据之间的关系。可追溯性使审阅者能够完成以下任务:
• Understand the construction of analysis datasets
• Determine the observations and algorithm(s) used to derive variables
• Understand how the confidence interval or the p-value was calculated in a particular analysis
• Relate counts from tables, listings, and figures in a study report to the underlying data
• 了解分析数据集的构造
• 确定用于派生变量的观测值和算法
• 了解特定分析中置信区间或 p 值的计算方式
• 将研究报告中的表、列表和数字的计数与基础数据相关联
Based upon reviewer experience, establishing traceability is one of the most problematic issues associated with any data conversion. If the reviewer is unable to trace study data from the data collection of subjects participating in a study to the analysis of the overall study data, then the regulatory review of a submission may be compromised. Traceability can be enhanced when studies are prospectively designed to collect data using a standardized CRF, e.g., CDASH. Traceability can be further enhanced when a flow diagram is submitted showing how data move from collection through preparation and submission to the Agency.
根据审阅者的经验,建立可追溯性是与任何数据转换相关的最成问题的问题之一。如果评价员无法追踪从参与研究的受试者的数据收集到对整体研究数据的分析的研究数据,那么对提交的监管审查可能会受到影响。当研究被前瞻性地设计为使用标准化的CRF(例如CDASH)收集数据时,可以增强可追溯性。提交流程图,说明数据如何从收集到准备和提交给机构,可以进一步提高可追溯性。
Reviewers evaluating nonclinical studies have similar needs to the above list, though in the case of nonclinical studies traceability allows the reviewer to understand and trace relationships between analysis results, single animal listings in the Study Report, and the tabulation data sets. Traceability between the Study Report and tabulation data can be enhanced when data in collection systems has a well-defined relationship to the SEND standard.
评估非临床研究的审稿人与上述列表有类似的需求,但在非临床研究的情况下,可追溯性允许审稿人理解和追踪分析结果、研究报告中的单个动物列表和制表数据集之间的关系。当收集系统中的数据与SEND标准具有明确定义的关系时,可以增强研究报告和制表数据之间的可追溯性。
8.3.2 Legacy Study Data Conversion to Standardized Study Data 遗留研究数据转换为标准化研究数据
Legacy study data are study data in a non-standardized format, not supported by FDA, and not ever listed in the Catalog. Sponsors should use processes for legacy data conversion that account for traceability. Generally, a conversion to a standard format will map every data element as originally collected to a corresponding data element described in a standard. Some study data conversions will be straightforward and will result in all data converted to a standardized format. In some instances, it may not be possible to represent a collected data element as a standardized data element. In these cases, there should be an explanation in the RG as to why certain data elements could not be fully standardized or were otherwise not included in the standardized data submission. The legacy data (i.e., aCRF, legacy tabulation data, and legacy analysis data) may be needed in addition to the submission of converted data.
遗留研究数据是非标准化格式的研究数据,不受FDA支持,也从未在目录中列出。申办者应使用考虑可追溯性的旧数据转换流程。通常,转换为标准格式会将最初收集的每个数据元素映射到标准中描述的相应数据元素。一些研究数据转换将很简单,并且将导致所有数据转换为标准化格式。在某些情况下,可能无法将收集的数据元素表示为标准化数据元素。在这些情况下,应在RG中解释为什么某些数据要素不能完全标准化,或者没有包括在标准化数据提交中。除了提交转换后的数据外,可能还需要遗留数据(即 aCRF、遗留制表数据和遗留分析数据)
In cases where the data were collected on a Case Report Form (CRF) or electronic CRF but were not included in the converted datasets, the omitted data should be apparent on the annotated CRF and described in the RG. The tabular list of studies in the Standardization Plan should indicate which studies contained previously collected non-standard data that were subsequently converted to a standard format.
如果数据是在病例报告表或电子通用报告格式上收集的,但未包括在转换后的数据集中,则省略的数据应在附加说明的通用报告格式上明显,并在RG中加以说明。标准化计划中的研究表格列表应指出哪些研究包含以前收集的非标准数据,这些数据随后被转换为标准格式。
For nonclinical studies where data are converted to SEND from a previously established collection system, instances may arise where it is not possible to represent a collected data element as a standardized data element. In these cases, there should be an explanation in the nSDRG as to why certain data elements could not be fully standardized or were otherwise not included in the standardized data submission. As the Study Report should contain a complete representation of the study data in the individual animal listings, no non-standardized electronic study data should be submitted.
对于从先前建立的收集系统将数据转换为SEND的非临床研究,可能会出现无法将收集的数据元素表示为标准化数据元素的情况。在这些情况下,nSDRG中应解释为什么某些数据要素不能完全标准化,或者没有包括在标准化数据提交中。由于研究报告应包含单个动物清单中研究数据的完整表示,因此不应提交非标准化的电子研究数据。
8.3.2.1 Traceability Issues with Legacy Data Conversion 遗留数据转换的可追溯性问题
FDA does not recommend a particular approach to legacy clinical study data conversion, but rather explains the issues that should be addressed so that the converted data are traceable and adequate to support review.
FDA不建议使用特定方法进行遗留临床研究数据转换,而是解释了应解决的问题,以便转换后的数据可追溯并足以支持审查。
Table 3 presents some of the issues that can be observed during a review when legacy study data are converted to SDTM and submitted with legacy analysis datasets.
表3列出了在将遗留研究数据转换为SDTM并与遗留分析数据集一起提交时,在审查期间可以观察到的一些问题。
 Table 4 presents the issues when legacy study data and legacy analysis data are independently converted to SDTM and ADaM formats, respectively, rather than ADaM datasets being created directly from the SDTM datasets (converted from legacy study data).
Table 4 presents the issues when legacy study data and legacy analysis data are independently converted to SDTM and ADaM formats, respectively, rather than ADaM datasets being created directly from the SDTM datasets (converted from legacy study data).
表 4 介绍了将遗留研究数据和遗留分析数据分别独立转换为 SDTM 和 ADaM 格式,而不是直接从 SDTM 数据集(从旧版研究数据转换)创建 ADaM 数据集时的问题。
Table 5 presents the issues when legacy data are converted to SDTM and ADaM formats in sequence (i.e., converting legacy study data to SDTM and then creating ADaM from the SDTM). The key concern is the traceability from ADaM to the Tables, Figures and CSR.
表 5 介绍了将旧数据按顺序转换为 SDTM 和 ADaM 格式时的问题(即,将旧研究数据转换为 SDTM,然后从 SDTM 创建 ADaM)。关键问题是从 ADaM 到表格、数字和 CSR 的可追溯性。
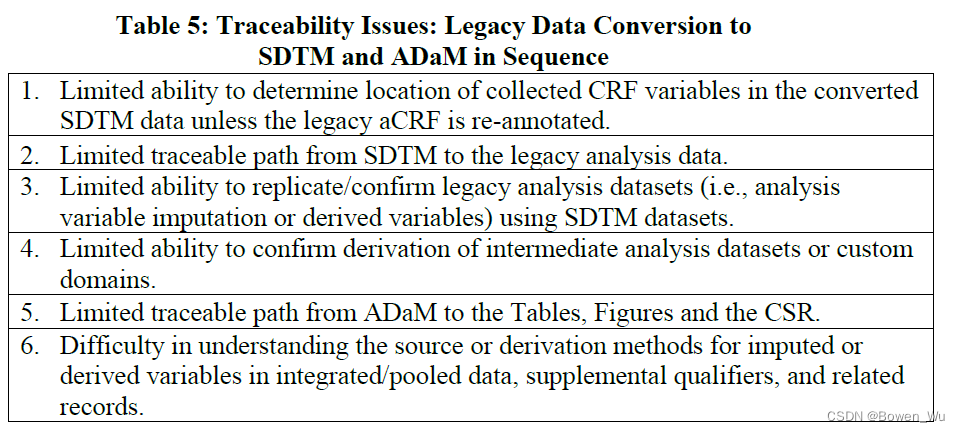
8.3.2.2 Legacy Data Conversion Plan and Report 遗留数据转换计划和报告
Sponsors should evaluate the decision involved in converting previously collected non-standardized data (i.e., legacy study data) to standardized data (i.e., SDTM, and ADaM). Sponsors should provide the explanation and rationale for the study data conversion in the RG. To mitigate traceability issues when converting legacy data, FDA recommends the following procedures:
申办者应评估将以前收集的非标准化数据(即遗留研究数据)转换为标准化数据(即SDTM和ADaM)所涉及的决策。申办者应在RG中提供研究数据转换的解释和理由。为了缓解转换遗留数据时的可追溯性问题,FDA建议采取以下程序:
1. Prepare and submit a legacy data conversion plan and report.
• The plan should describe the legacy data and the process intended for the conversion.
• The report should present the results of the conversions, issues encountered and resolved, and outstanding issues.
• The plan and report should be provided in the SDRG.
1. 准备并提交旧数据转换计划和报告。
• 计划应描述旧数据和用于转换的过程。
• 报告应说明转换的结果、遇到和解决的问题以及悬而未决的问题。
• 计划和报告应在SDRG中提供。
2. Provide an aCRF, for clinical data, that maps the legacy data elements.
• Sponsors should provide two separate CRF annotations, one based on the original legacy data, and the other based on the converted data (i.e., SDTM) when legacy datasets are submitted. The legacy CRF tabulation data should include all versions and all forms used in the study.
• Record significant data issues, clarifications, explanations of traceability, and adjudications in the RG. For example, data were not collected or were collected using different/incompatible terminologies, or were collected but will not fit into, for example, SDTM format.
• Legacy data (i.e., legacy aCRF, legacy tabulation data, and legacy analysis data) may be needed in addition to the converted data.
2. 为临床数据提供 aCRF,以映射遗留数据元素。
•申办者应提供两个单独的CRF注释,一个基于原始遗留数据,另一个基于提交遗留数据集时转换后的数据(即SDTM)。传统的通用报告格式制表数据应包括研究中使用的所有版本和所有表格。
•在RG中记录重要的数据问题,澄清,可追溯性解释和裁决。例如,未收集数据或使用不同/不兼容的术语收集数据,或者收集数据但不适合 SDTM 格式等。
• 除了转换后的数据外,可能还需要遗留数据(即遗留 aCRF、遗留制表数据和遗留分析数据)。
Submission of a Legacy Data Conversion Plan and Report is not expected for nonclinical studies where data were collected in a previously established data collection system.
对于在先前建立的数据收集系统中收集数据的非临床研究,预计不会提交遗留数据转换计划和报告。
Appendix A: Data Standards and Interoperable Data Exchange 附录 A:数据标准和可互操作的数据交换
[译注:略]
Appendix B: Trial Summary (TS) Parameters for Submission – Clinical 附录 B:提交试验摘要 (TS) 参数 – 临床
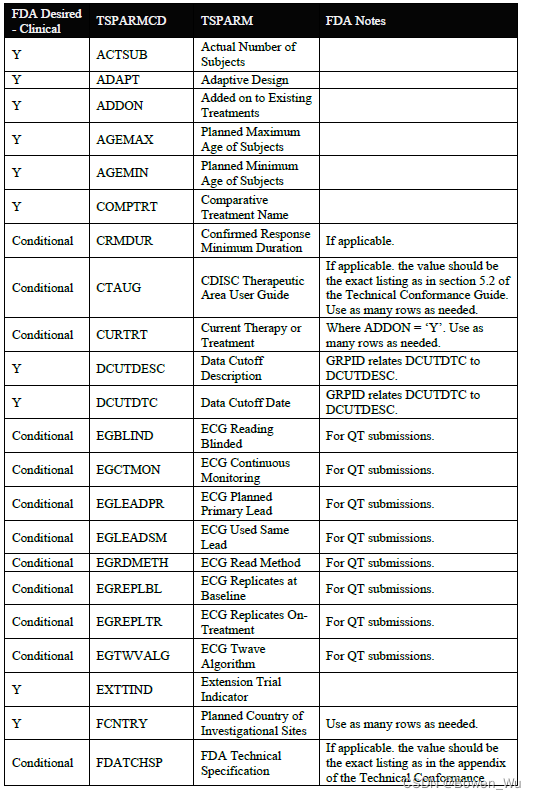
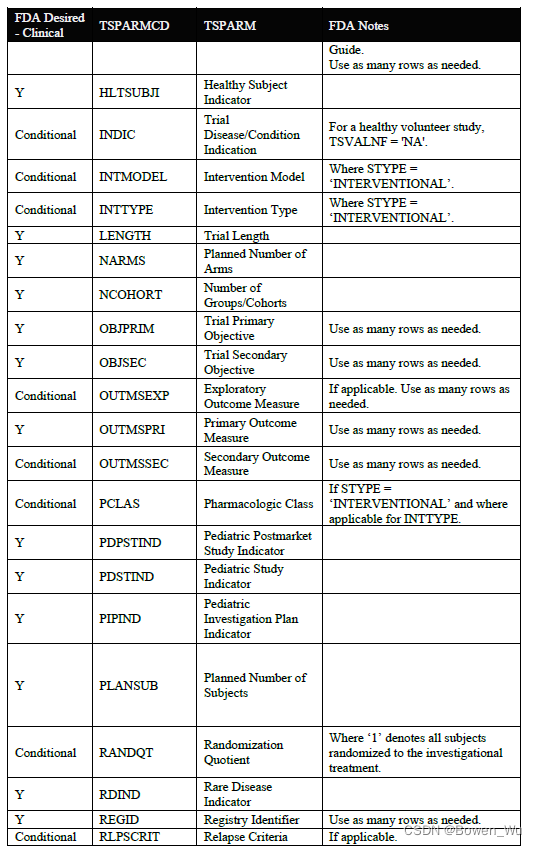
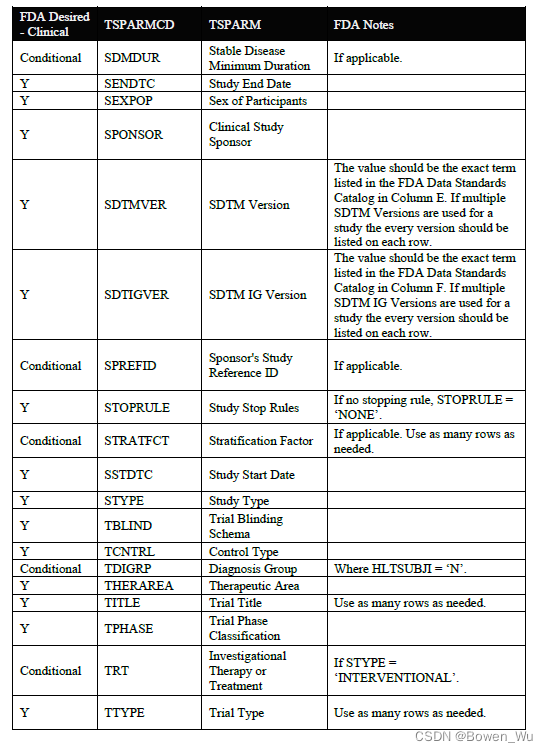
Appendix C: Trial Summary (TS) Parameters for Submission – Nonclinical 附录 C:提交试验摘要 (TS) 参数 – 非临床
[译注:略]
Appendix D: Additional Documents Evaluated By FDA 附录D:FDA评估的其他文件
The Agency recognizes that there are may be additional documents beyond Therapeutic Area User Guides (TAUGs), Implementation Guides (IGs), and Models that provide technical information about how to implement a CDISC standard and that these documents fall outside the scope of the FDA Data Standards Catalog. Use of the documents listed here is encouraged. For documents not yet listed here, please consult with your division.
该机构认识到,除了治疗领域用户指南(TAUGs),实施指南(IG)和模型之外,可能还有其他文档,这些文件提供了有关如何实施CDISC标准的技术信息,并且这些文件不属于FDA数据标准目录的范围。鼓励使用此处列出的文档。对于此处尚未列出的文件,请咨询您的部门。
1. CDISC Document: Interim User Guide for COVID-19 CDISC 文档:COVID-19 临时用户指南
2. CDISC Document: Guidance for Ongoing Studies Disrupted by COVID-19 Pandemic CDISC文件:受COVID-19大流行干扰的持续研究指南
It is the current preference of the Agency that for all clinical studies, not limited to those impacted by COVID-19, subject visit data for scheduled (whether or not they occurred), and unscheduled visits be submitted in one single dataset structured as the current CDISC Subject Visits (SV) domain. It is also Agency preference that three non-standard variables (NSVs) for missed visits, --REASOC (Reason for Occur Value), --EPCHGI (Epi/Pandemic Related Change Indicator), and --CNTMOD (Contact Mode), outlined in the CDISC document “Guidance for Ongoing Studies Disrupted by COVID-19 Pandemic” be included within the SV domain and not within the supplemental SUPPSV domain or in other SDTM datasets. Submitting subject visits information in one single structured dataset allows both the human and technology consumer of this information to operate efficiently and with confidence that all visit data are considered during regulatory review.
机构目前倾向于为所有临床研究(不限于受COVID-19影响的研究),计划(无论是否发生)的受试者就诊数据以及计划外就诊提交到一个单一数据集中,该数据集的结构为当前的CDISC受试者就诊(SV)域。机构还倾向于将CDISC文件“COVID-19大流行中断的持续研究指南”中概述的三个非标准变量(NSV)包含在SV域内,而不是包含在补充SUPSV域或其他SDPSV数据集中。在单个结构化数据集中提交受试者访问信息,使此信息的人类和技术消费者能够高效运行,并确信在监管审查期间会考虑所有访问数据。
As always, consult with the relevant FDA review division for the best approach in a specific application. Further updates to Agency thinking regarding how to submit data for studies that may have been impacted by the COVID-19 pandemic will be posted in updates to the Study Data Technical Conformance Guide.
与往常一样,请咨询相关的FDA审查部门,了解特定应用中的最佳方法。机构关于如何为可能受COVID-19大流行影响的研究提交数据的想法的进一步更新将在研究数据技术一致性指南的更新中发布。
3. Occurrence Dataset Structure (OCCDS) v1.0
Appendix E: Example Study Data Folder Structure 附录 E:示例算例研究数据文件夹结构
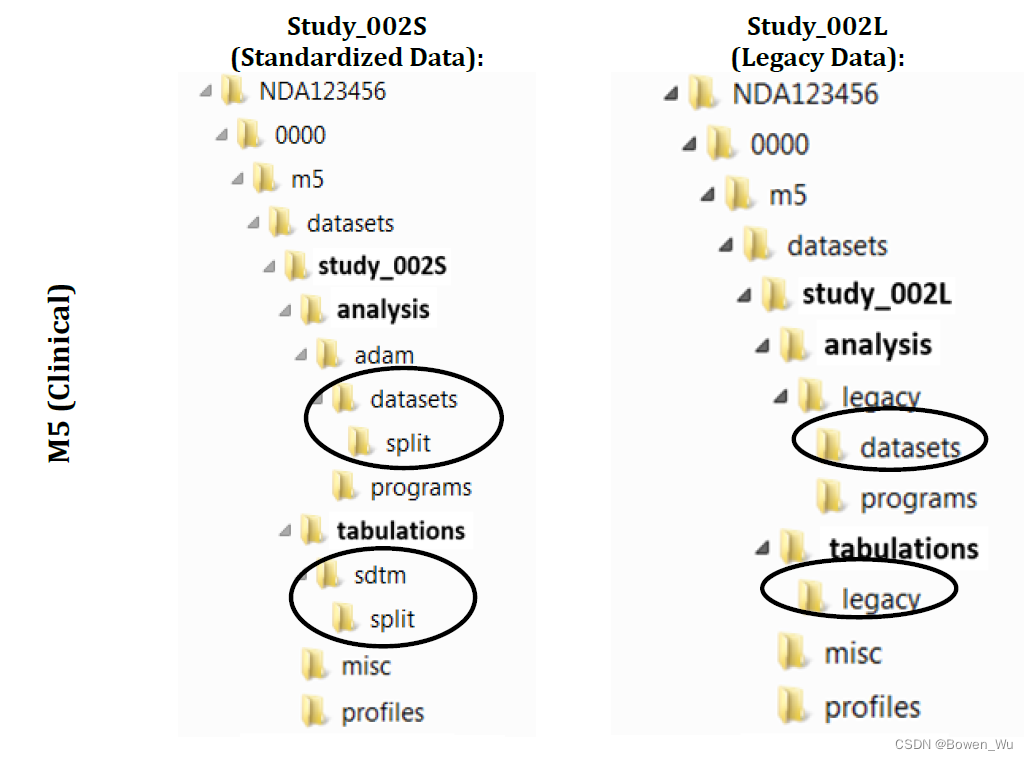
Appendix F/G 略