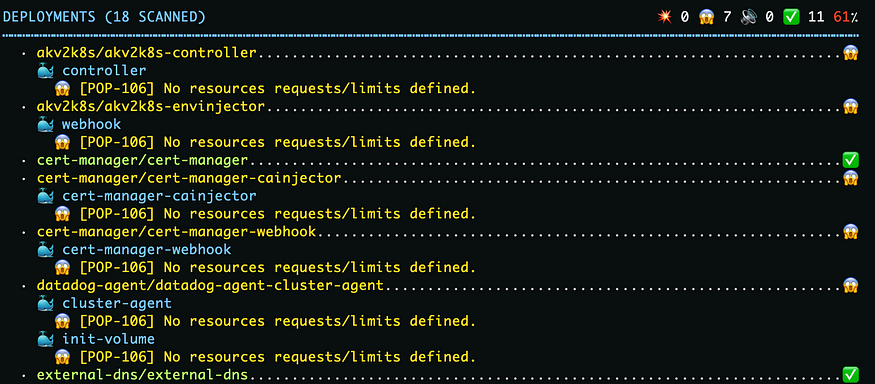
📋文章目录
- 面积图简介
- 准备数据集
- 加载数据集
- 数据处理
- 数据可视化
利用R语言绘制物种组成图。本文以堆叠面积图的方式与大家分享。
面积图简介
面积图又叫区域图。它是在折线图的基础之上形成的, 它将折线图中折线与自变量坐标轴之间的区域使用颜色或者纹理填充,这样一个填充区域我们叫面积。颜色的填充可以更好地突出趋势信息(比如时间上的差异,分类上的差异),需要注意的是颜色要带有一定的透明度,透明度可以很好地帮助使用者观察不同序列之间的重叠关系,没有透明度的面积会导致不同序列之间相互遮盖减少可以被观察到的信息。
与折线图相似,面积图可用于强调数量随时间或分类而变化的程度,也可用于引起人们对总值趋势的注意。他们最常用于表现趋势和关系,而不是传达特定的值。
- 所有的数据都从相同的零轴开始。
- 标准面积图适用于展示或者比较随着时间连续变化的定量。
- 在需要绘制大量数据系列的情况下,折线图通常是更清晰的可视化表达方式。

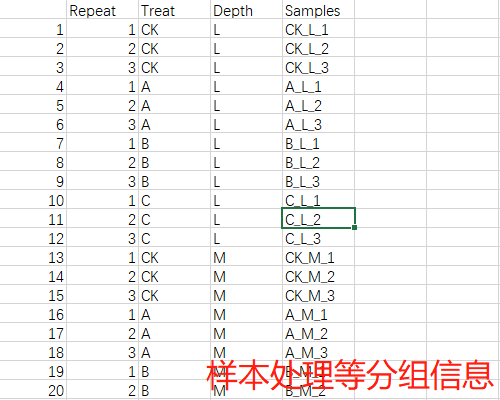
准备数据集


加载数据集
otudf <- read.csv("otudf.csv", header = T, row.names = 1)
group_infor <- read.csv("group_infor.csv", header = T, row.names = 1)head(otudf[1:6, 1:6])
# CK_L_1 CK_L_2 CK_L_3 A_L_1 A_L_2 A_L_3
# otu_1 414 1371 113 1190 427 1327
# otu_2 462 1028 937 394 1266 547
# otu_3 178 193 722 1421 705 579
# otu_4 525 56 822 885 690 944
# otu_5 194 1353 365 680 1218 677
# otu_6 937 1140 1499 1098 1460 494
head(group_infor)
# Repeat Treat Depth Samples
# 1 1 CK L CK_L_1
# 2 2 CK L CK_L_2
# 3 3 CK L CK_L_3
# 4 1 A L A_L_1
# 5 2 A L A_L_2
# 6 3 A L A_L_3
数据处理
## 提取物种丰度和物种注释信息
species <- otudf[c(1:36)]
taxa <- otudf[-c(1:36)]## 确定数据正确与否
head(species)
# CK_L_1 CK_L_2 CK_L_3 A_L_1 A_L_2 A_L_3 B_L_1 B_L_2 B_L_3 C_L_1 C_L_2 C_L_3 CK_M_1 CK_M_2 CK_M_3 A_M_1 A_M_2 A_M_3
# otu_1 414 1371 113 1190 427 1327 631 207 1473 1322 1370 1476 28 1271 125 978 945 0
# otu_2 462 1028 937 394 1266 547 1144 851 365 1418 651 1329 1061 1124 332 919 1483 600
# otu_3 178 193 722 1421 705 579 1103 1398 241 1328 1286 452 263 1446 893 1098 868 661
# otu_4 525 56 822 885 690 944 86 720 1135 37 1323 792 362 696 806 1398 11 37
# otu_5 194 1353 365 680 1218 677 1129 547 829 1038 103 276 1166 630 539 1298 277 663
# otu_6 937 1140 1499 1098 1460 494 988 709 1137 251 356 130 344 727 1332 516 1154 454
# B_M_1 B_M_2 B_M_3 C_M_1 C_M_2 C_M_3 CK_H_1 CK_H_2 CK_H_3 A_H_1 A_H_2 A_H_3 B_H_1 B_H_2 B_H_3 C_H_1 C_H_2 C_H_3
# otu_1 1394 213 1449 1431 786 697 627 463 935 58 118 967 864 226 1053 834 551 961
# otu_2 985 247 1135 943 584 264 1286 838 37 1057 320 982 1351 621 1058 667 696 780
# otu_3 1117 566 508 142 1016 924 244 1426 1093 994 1166 962 632 628 163 1455 756 878
# otu_4 244 787 1261 533 1025 583 201 533 1051 651 1294 20 206 1151 369 1499 358 1083
# otu_5 716 736 1219 1411 1295 3 434 1023 406 1215 1074 994 1220 1220 528 1349 1138 906
# otu_6 1053 163 295 412 473 317 1065 981 1036 1113 435 19 1396 1151 1048 134 1199 383
head(taxa)
# phylum class order family genus species
# otu_1 Spirochaetes Gemmatimonadetes BRC1 Firmicutes Planctomycetes Nitrospirae
# otu_2 WPS_1 Euryarchaeota Cyanobacteria Armatimonadetes Spirochaetes Chloroflexi
# otu_3 Gemmatimonadetes Gemmatimonadetes Armatimonadetes Cyanobacteria WPS_2 WPS_1
# otu_4 WPS_1 Chloroflexi Acidobacteria Firmicutes Acidobacteria WPS_2
# otu_5 Acidobacteria Cyanobacteria BRC1 Chloroflexi Bacteroidetes Spirochaetes
# otu_6 Planctomycetes Gemmatimonadetes WPS_1 Chloroflexi Chloroflexi Armatimonadetes# 计算不同处理下的门水平均值
library(tidyverse)
Phylum <- species %>%group_by(taxa$phylum) %>% # 使用taxa中的门水平进行分类summarise_all(sum) %>% # 计算总的otu数量rename(Phylum = `taxa$phylum`) %>% # 修改名称gather(key = "Samples", value = "Abundance", -Phylum) %>% # 数据形式转换 宽-长left_join(group_infor, by = c("Samples" = "Samples")) %>%select(Phylum, Treat, Depth, Abundance) %>%group_by(Phylum, Treat, Depth) %>% # 求均值summarise_all(mean) %>%arrange(Phylum, Treat, Depth, desc(Abundance))
Phylum
# # A tibble: 204 × 4
# # Groups: Phylum, Treat [68]
# Phylum Treat Depth Abundance
# <chr> <chr> <chr> <dbl>
# 1 Acidobacteria A H 462665.
# 2 Acidobacteria A L 459384.
# 3 Acidobacteria A M 454049.
# 4 Acidobacteria B H 454310
# 5 Acidobacteria B L 455244.
# 6 Acidobacteria B M 448743.
# 7 Acidobacteria C H 449425
# 8 Acidobacteria C L 455059.
# 9 Acidobacteria C M 454707.
# 10 Acidobacteria CK H 456931
# # … with 194 more rows
# # ℹ Use `print(n = ...)` to see more rows## 选择top9 及 合并剩余物种作为Other
phy_select <- unique(Phylum$Phylum)[1:9]top9 <- Phylum[Phylum$Phylum %in% phy_select, ]
other <- Phylum[!Phylum$Phylum %in% phy_select, ]
other <- other %>% group_by(Treat, Depth) %>% summarise(Abundance = mean(Abundance)) %>% cbind(Phylum = "Other") %>%select(Phylum, Treat, Depth, Abundance, everything())top10 <- rbind(top9, other)
head(top10)
# # A tibble: 6 × 4
# # Groups: Phylum, Treat [2]
# Phylum Treat Depth Abundance
# <chr> <chr> <chr> <dbl>
# 1 Acidobacteria A H 462665.
# 2 Acidobacteria A L 459384.
# 3 Acidobacteria A M 454049.
# 4 Acidobacteria B H 454310
# 5 Acidobacteria B L 455244.
# 6 Acidobacteria B M 448743.
数据可视化
# 物种组成堆叠面积图
library(ggplot2)
library(ggalluvial)
ggplot(data = top10,aes(x = Depth, y = Abundance, fill = reorder(Phylum, -Abundance),colour = reorder(Phylum, -Abundance),stratum = reorder(Phylum, -Abundance) ,alluvium = reorder(Phylum, -Abundance))) +geom_alluvium(aes(fill = reorder(Phylum, -Abundance)), alpha = 0.7, decreasing = FALSE) +geom_stratum(aes(fill = reorder(Phylum, Abundance)), width = 0.3, size = 0.1, color = "black") +scale_y_continuous(expand = c(0, 0)) +theme_bw() +facet_grid(. ~ Treat, scales = "fixed") +scale_fill_manual(values = c("#EB7369", "#CF8B0B", "#9D9F20", "#2BB077", "#2BB077","#1BB3B7", "#29A4DE", "#8989C1", "#B174AD","#DE66A1"), name = "Phylum") +scale_color_manual(values = c("#EB7369", "#CF8B0B", "#9D9F20", "#2BB077", "#2BB077","#1BB3B7", "#29A4DE", "#8989C1", "#B174AD","#DE66A1")) +guides(color = "none")+theme(panel.grid=element_blank(),panel.spacing.x = unit(0, units = "cm"),strip.background = element_rect(color = "white", fill = "white", linetype = "solid", size = 1),strip.placement = "outside",axis.line.y.left = element_line(color = "black", size = 0.7),axis.line.x.bottom = element_line(color = "black", size = 0.7),strip.text.x = element_text(size = 14, face = "bold"),axis.text = element_text(face = "bold", size = 12, color = "black"),axis.title = element_text(face = "bold", size = 14, colour = "black"),legend.title = element_text(face = "bold", size = 12, color = "black"),legend.text = element_text(face = "bold", size = 12, color = "black"),axis.ticks.x = element_line(size = 1),axis.ticks.y = element_line(size = 1),)+labs(x = "Depth",y= "Relative Abundance of Phylum (%)")