使用U-Net实现肾脏CT分割
- 数据集准备
- 数据来源
- 数据预处理
- 网络结构及代码
- 网络结构
- 训练代码
- 训练过程
- 参数设置:
- 可视化
- 结果分析
数据集准备
数据来源
MICCAI KiTS19(Kidney Tumor Segmentation Challenge):https://kits19.grand-challenge.org/
KiTS2019是MICCAI19的一个竞赛项目,项目的任务是对3D-CT数据进行肾脏和肾脏肿瘤的分割,官方的数据集提供了210个case作为训练集,90个case作为测试集。共有800多人报名参加了这一竞赛,最终提交的结果的team有126支,其中被认定有效的为100个记录入leaderboard。目前这一竞赛状态为开放性质的,有兴趣的可以参与一下。这一挑战将于2021年继续举办,将提供更多的数据集和标注,任务也将变得更加有挑战,有兴趣的同学可以跟进关注一下。
感谢评论里weixin_40621562老哥提供的数据集百度云版:链接: https://pan.baidu.com/s/1AOQDjPz9ye32DH-oDS0WDw 提取码: d7jk
数据预处理
KiTS19提供的数据是3D CT图像,我们要训练的是最简单的2D U-Net,因此要从3D CT体数据中读取2D切片。数据集的提供方在其Github上很贴心的提供了可视化的代码,是用python调用了nibabel库处理.nii格式的体数据得到2D的.png格式的切片。可视化的结果如下图所示,需要对切片进行筛选。另外需要补充的是在KiTS的数据集中分割的标签有三类:背景、肾脏、肾脏肿瘤,我们想进行的是简单的背景与肾脏二分类问题而不是多分类问题,因此在可视化过程中比较简单粗暴的将肿瘤视为肾脏的一部分。
一共有210个3D CT的体数据,从每个体数据中选取了10个slice,一共得到了2100张2D的.png格式图像。

网络结构及代码
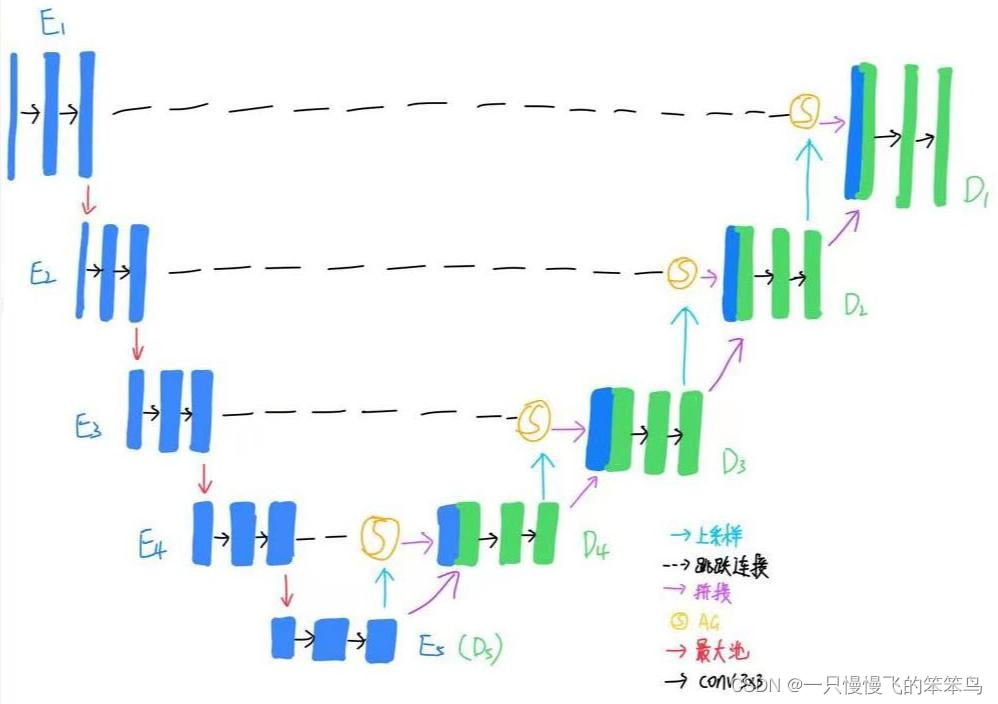
网络结构
U-Net的结构时最简单的encoder-decoder结构,再加上越级连接,详细的网络结构请见我的另一篇博客深度学习图像语义分割网络总结:U-Net与V-Net的Pytorch实现。
训练代码
代码参照了github上的https://github.com/milesial/Pytorch-UNet
import argparse
import logging
import os
import sysimport numpy as np
import torch
import torch.nn as nn
from torch import optim
from tqdm import tqdmfrom eval import eval_net
from unet import UNetfrom visdom import Visdom
from utils.dataset import BasicDataset
from torch.utils.data import DataLoader, random_splitdir_img = 'D:\Dataset\CT-KiTS19\KiTS19\kits19-master\png_datasize\\train_choose\slice_png'
dir_mask = 'D:\Dataset\CT-KiTS19\KiTS19\kits19-master\png_datasize\\train_choose\mask_png'
dir_checkpoint = 'checkpoints/'def train_net(net,device,epochs=5,batch_size=1,lr=0.1,val_percent=0.2,save_cp=True,img_scale=1):dataset = BasicDataset(dir_img, dir_mask, img_scale)n_val = int(len(dataset) * val_percent)n_train = len(dataset) - n_valtrain, val = random_split(dataset, [n_train, n_val])train_loader = DataLoader(train, batch_size=batch_size, shuffle=True, num_workers=4, pin_memory=True)val_loader = DataLoader(val, batch_size=batch_size, shuffle=False, num_workers=4, pin_memory=True, drop_last=True)#writer = SummaryWriter(comment=f'LR_{lr}_BS_{batch_size}_SCALE_{img_scale}')viz=Visdom()viz.line([0.], [0.], win='train_loss', opts=dict(title='train_loss'))viz.line([0.], [0.], win='learning_rate', opts=dict(title='learning_rate'))viz.line([0.], [0.], win='Dice/test', opts=dict(title='Dice/test'))global_step = 0logging.info(f'''Starting training:Epochs: {epochs}Batch size: {batch_size}Learning rate: {lr}Training size: {n_train}Validation size: {n_val}Checkpoints: {save_cp}Device: {device.type}Images scaling: {img_scale}''')optimizer = optim.RMSprop(net.parameters(), lr=lr, weight_decay=1e-8, momentum=0.9)scheduler = optim.lr_scheduler.ReduceLROnPlateau(optimizer, 'min' if net.n_classes > 1 else 'max', patience=2)if net.n_classes > 1:criterion = nn.CrossEntropyLoss()else:criterion = nn.BCEWithLogitsLoss()for epoch in range(epochs):net.train()epoch_loss = 0with tqdm(total=n_train, desc=f'Epoch {epoch + 1}/{epochs}', unit='img') as pbar:for batch in train_loader:imgs = batch['image']true_masks = batch['mask']assert imgs.shape[1] == net.n_channels, \f'Network has been defined with {net.n_channels} input channels, ' \f'but loaded images have {imgs.shape[1]} channels. Please check that ' \'the images are loaded correctly.'imgs = imgs.to(device=device, dtype=torch.float32)mask_type = torch.float32 if net.n_classes == 1 else torch.longtrue_masks = true_masks.to(device=device, dtype=mask_type)masks_pred = net(imgs)#print('mask_pred',masks_pred.shape)#print('masks_pred',masks_pred.shape)#print('true_masks', true_masks.shape)viz.image(imgs, win='imgs/train')viz.image(true_masks, win='masks/true/train')viz.image(masks_pred, win='masks/pred/train')loss = criterion(masks_pred, true_masks)epoch_loss += loss.item()#writer.add_scalar('Loss/train', loss.item(), global_step)viz.line([loss.item()],[global_step],win='train_loss',update='append')pbar.set_postfix(**{'loss (batch)': loss.item()})optimizer.zero_grad()loss.backward()#nn.utils.clip_grad_value_(net.parameters(), 0.1)optimizer.step()pbar.update(imgs.shape[0])global_step += 1if global_step % (n_train // (10 * batch_size)) == 0:# for tag, value in net.named_parameters():# tag = tag.replace('.', '/')# writer.add_histogram('weights/' + tag, value.data.cpu().numpy(), global_step)# writer.add_histogram('grads/' + tag, value.grad.data.cpu().numpy(), global_step)val_score = eval_net(net, val_loader, device)scheduler.step(val_score)#writer.add_scalar('learning_rate', optimizer.param_groups[0]['lr'], global_step)viz.line([optimizer.param_groups[0]['lr']], [global_step], win='learning_rate', update='append')if net.n_classes > 1:logging.info('Validation cross entropy: {}'.format(val_score))#writer.add_scalar('Loss/test', val_score, global_step)else:logging.info('Validation Dice Coeff: {}'.format(val_score))#writer.add_scalar('Dice/test', val_score, global_step)viz.line([val_score], [global_step], win='Dice/test', update='append')viz.image(imgs, win='images')if net.n_classes == 1:print('true_mask',true_masks.shape,true_masks.type)viz.image( true_masks, win='masks/true')print('pred',(torch.sigmoid(masks_pred) > 0.5).squeeze(0).shape)viz.images((torch.sigmoid(masks_pred) > 0.5),win='masks/pred')if save_cp:try:os.mkdir(dir_checkpoint)logging.info('Created checkpoint directory')except OSError:passtorch.save(net.state_dict(),dir_checkpoint + f'CP_epoch{epoch + 1}.pth')logging.info(f'Checkpoint {epoch + 1} saved !')#writer.close()def eval_net(net, loader, device):"""Evaluation without the densecrf with the dice coefficient"""net.eval()mask_type = torch.float32 #if net.n_classes == 1 else torch.longn_val = len(loader) # the number of batchtot = 0with tqdm(total=n_val, desc='Validation round', unit='batch', leave=False) as pbar:for batch in loader:imgs, true_masks = batch['image'], batch['mask']imgs = imgs.to(device=device, dtype=torch.float32)true_masks = true_masks.to(device=device, dtype=mask_type)with torch.no_grad():mask_pred = net(imgs)#['out']# if net.n_classes > 1:# tot += F.cross_entropy(mask_pred, true_masks).item()# else:pred = torch.sigmoid(mask_pred)pred = (pred > 0.5).float()tot += dice_coeff(pred, true_masks).item()pbar.update()net.train()return tot / n_valdef get_args():parser = argparse.ArgumentParser(description='Train the UNet on images and target masks',formatter_class=argparse.ArgumentDefaultsHelpFormatter)parser.add_argument('-e', '--epochs', metavar='E', type=int, default=5,help='Number of epochs', dest='epochs')parser.add_argument('-b', '--batch-size', metavar='B', type=int, nargs='?', default=1,help='Batch size', dest='batchsize')parser.add_argument('-l', '--learning-rate', metavar='LR', type=float, nargs='?', default=0.1,help='Learning rate', dest='lr')parser.add_argument('-f', '--load', dest='load', type=str, default=False,help='Load model from a .pth file')parser.add_argument('-s', '--scale', dest='scale', type=float, default=1,help='Downscaling factor of the images')parser.add_argument('-v', '--validation', dest='val', type=float, default=20.0,help='Percent of the data that is used as validation (0-100)')return parser.parse_args()if __name__ == '__main__':logging.basicConfig(level=logging.INFO, format='%(levelname)s: %(message)s')args = get_args()device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')logging.info(f'Using device {device}')# Change here to adapt to your data# n_channels=3 for RGB images# n_classes is the number of probabilities you want to get per pixel# - For 1 class and background, use n_classes=1# - For 2 classes, use n_classes=1# - For N > 2 classes, use n_classes=Nnet = UNet(n_channels=1, n_classes=1, bilinear=True)logging.info(f'Network:\n'f'\t{net.n_channels} input channels\n'f'\t{net.n_classes} output channels (classes)\n'f'\t{"Bilinear" if net.bilinear else "Transposed conv"} upscaling')if args.load:net.load_state_dict(torch.load(args.load, map_location=device))logging.info(f'Model loaded from {args.load}')net.to(device=device)# faster convolutions, but more memory# cudnn.benchmark = Truetry:train_net(net=net,epochs=args.epochs,batch_size=args.batchsize,lr=args.lr,device=device,img_scale=args.scale,val_percent=args.val / 100)except KeyboardInterrupt:torch.save(net.state_dict(), 'INTERRUPTED.pth')logging.info('Saved interrupt')try:sys.exit(0)except SystemExit:os._exit(0)
训练过程
参数设置:
训练与验证比例: 8:2 (1680:420)
batch_size: 2
学习率:torch.optim.lr_scheduler.ReduceLROnPlateau,当网络的评价指标不在提升的时候,可以通过降低网络的学习率来提高网络性能损失函数:BCEWithLogitsLoss 衡量目标和输出之间的二进制交叉熵
可视化
使用visdom进行可视化
一开始的训练状态,左边为真实的mask,右边为网络的输出,可以看到一开始网络的输出还是不太行的。
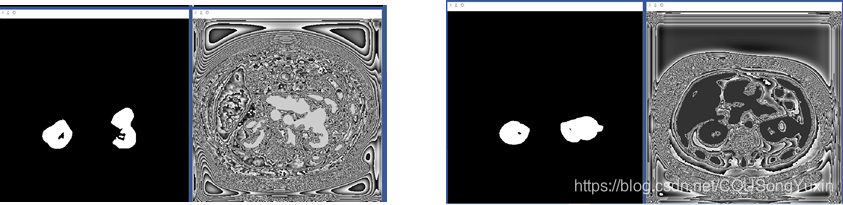
当进行完第一轮训练之后训练的结果如图所示,红色所框的为训练过程,蓝色所框为验证过程,包括了原图、真实的mask T、预测的mask P。训练和验证过程中预测mask的差异来自于是否进行了二值化处理。

第四轮训练之后的结果,预测的mask与真实的mask已经很接近了。

结果分析

实验结果:Dice系数:0.832
结果分析:
1.原数据为三维,本次实验只使用的二维切片
2.原数据的mask肿瘤和肾脏是分开的,在数据处理过程中统一化为了肾脏。
3.没有做数据增强、参数调整,训练不够充分。